rs4903712
Homo sapiens
T>C
NRXN3 : Intron Variant
SNV (Single Nucleotide Variation)
C=0194 (5808/29904,GnomAD)
C=0156 (4566/29118,TOPMED)
C=0129 (648/5008,1000G)
C=0276 (1063/3854,ALSPAC)
C=0266 (987/3708,TWINSUK)
C=0156 (4566/29118,TOPMED)
C=0129 (648/5008,1000G)
C=0276 (1063/3854,ALSPAC)
C=0266 (987/3708,TWINSUK)
chr14:78149250 (GRCh38.p7) (14q24.3)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 14 | NC_000014.9:g.78149250T>C |
| GRCh37.p13 chr 14 | NC_000014.8:g.78615593T>C |
Gene: NRXN3, neurexin 3(plus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| NRXN3 transcript variant 3 | NM_001105250.2:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant 4 | NM_001272020.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant 1 | NM_004796.5:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant 2 | NM_138970.4:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant 5 | NR_073546.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant 6 | NR_073547.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X2 | XM_011537364.2:c. | N/A | Intron Variant |
| NRXN3 transcript variant X3 | XM_017021790.1:c. | N/A | Intron Variant |
| NRXN3 transcript variant X4 | XM_017021791.1:c. | N/A | Intron Variant |
| NRXN3 transcript variant X6 | XM_005268218.2:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X7 | XM_006720322.2:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X21 | XM_006720323.2:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X1 | XM_011537363.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X5 | XM_011537365.2:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X9 | XM_011537366.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X15 | XM_011537367.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X19 | XM_011537368.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X20 | XM_011537369.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X21 | XM_011537370.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X25 | XM_011537371.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X26 | XM_011537372.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X31 | XM_011537373.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X44 | XM_011537377.2:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X8 | XM_017021792.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X10 | XM_017021793.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X11 | XM_017021794.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X12 | XM_017021795.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X13 | XM_017021796.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X13 | XM_017021797.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X14 | XM_017021798.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X17 | XM_017021799.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X22 | XM_017021800.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X23 | XM_017021801.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X24 | XM_017021802.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X27 | XM_017021803.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X28 | XM_017021804.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X29 | XM_017021805.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X30 | XM_017021806.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X30 | XM_017021807.1:c. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X32 | XR_001750599.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X33 | XR_001750600.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X34 | XR_001750601.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X35 | XR_001750602.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X36 | XR_001750603.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X37 | XR_001750604.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X38 | XR_001750605.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X39 | XR_001750606.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X40 | XR_001750607.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X41 | XR_001750608.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X42 | XR_001750609.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X44 | XR_001750610.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X45 | XR_943561.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X47 | XR_943562.1:n. | N/A | Genic Upstream Transcript Variant |
| NRXN3 transcript variant X46 | XR_943563.2:n. | N/A | Genic Upstream Transcript Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | T=0.965 | C=0.035 |
| 1000Genomes | American | Sub | 694 | T=0.820 | C=0.180 |
| 1000Genomes | East Asian | Sub | 1008 | T=0.985 | C=0.015 |
| 1000Genomes | Europe | Sub | 1006 | T=0.735 | C=0.265 |
| 1000Genomes | Global | Study-wide | 5008 | T=0.871 | C=0.129 |
| 1000Genomes | South Asian | Sub | 978 | T=0.800 | C=0.200 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | T=0.724 | C=0.276 |
| The Genome Aggregation Database | African | Sub | 8702 | T=0.936 | C=0.064 |
| The Genome Aggregation Database | American | Sub | 836 | T=0.810 | C=0.190 |
| The Genome Aggregation Database | East Asian | Sub | 1616 | T=0.994 | C=0.006 |
| The Genome Aggregation Database | Europe | Sub | 18448 | T=0.729 | C=0.270 |
| The Genome Aggregation Database | Global | Study-wide | 29904 | T=0.805 | C=0.194 |
| The Genome Aggregation Database | Other | Sub | 302 | T=0.680 | C=0.320 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | T=0.843 | C=0.156 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | T=0.734 | C=0.266 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 23691058 | Dosage transmission disequilibrium test (dTDT) for linkage and association detection. | Zhang Z | PLoS One |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs4903712 | 1.67E-06 | alcohol dependence | 23691058 |
eQTL of rs4903712 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs4903712 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
Genomic View
Chromatin Interaction
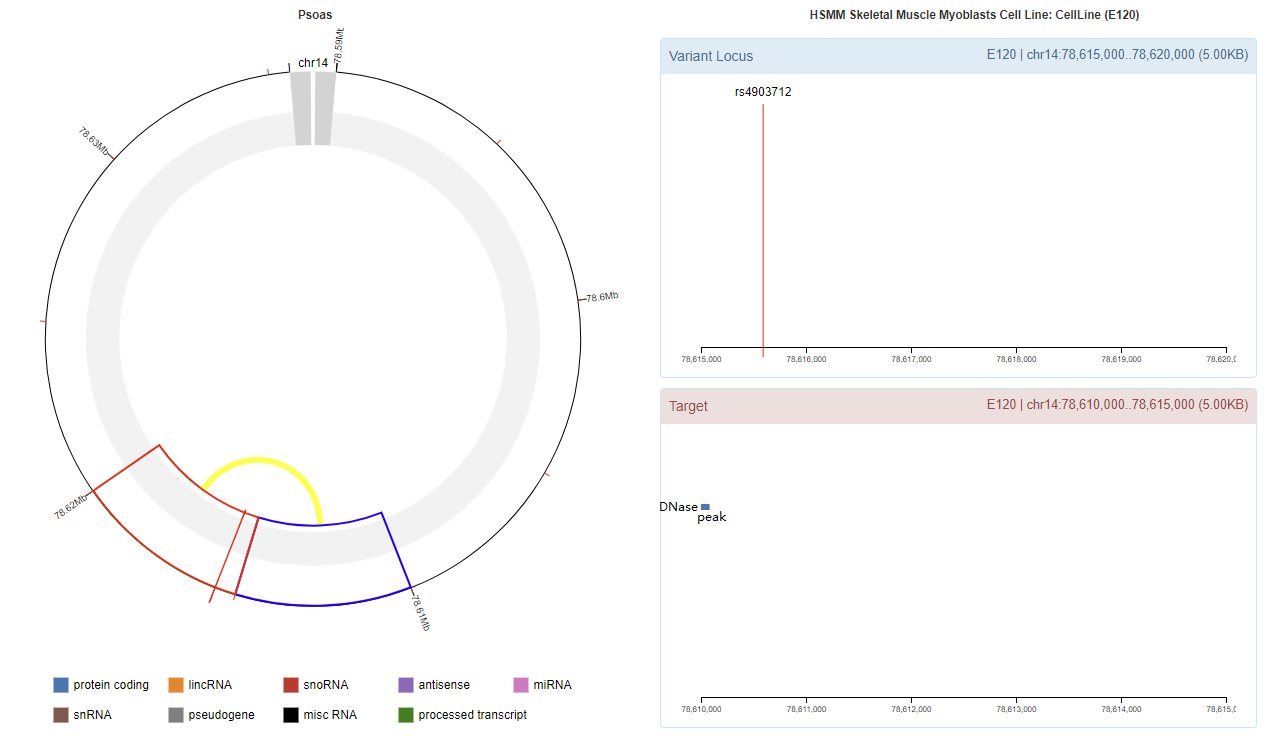
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr14 | 78597716 | 78597815 | E067 | -17778 |
| chr14 | 78598089 | 78598345 | E067 | -17248 |
| chr14 | 78598550 | 78598647 | E067 | -16946 |
| chr14 | 78636028 | 78636078 | E067 | 20435 |
| chr14 | 78636126 | 78636179 | E067 | 20533 |
| chr14 | 78644175 | 78644711 | E067 | 28582 |
| chr14 | 78644827 | 78644918 | E067 | 29234 |
| chr14 | 78598089 | 78598345 | E068 | -17248 |
| chr14 | 78598550 | 78598647 | E068 | -16946 |
| chr14 | 78597716 | 78597815 | E069 | -17778 |
| chr14 | 78598089 | 78598345 | E069 | -17248 |
| chr14 | 78618594 | 78619202 | E069 | 3001 |
| chr14 | 78588944 | 78589309 | E070 | -26284 |
| chr14 | 78589357 | 78589425 | E070 | -26168 |
| chr14 | 78619251 | 78619349 | E070 | 3658 |
| chr14 | 78619421 | 78619559 | E070 | 3828 |
| chr14 | 78619643 | 78619835 | E070 | 4050 |
| chr14 | 78623725 | 78623775 | E070 | 8132 |
| chr14 | 78623948 | 78623998 | E070 | 8355 |
| chr14 | 78624022 | 78624090 | E070 | 8429 |
| chr14 | 78624329 | 78624389 | E070 | 8736 |
| chr14 | 78624432 | 78624724 | E070 | 8839 |
| chr14 | 78642325 | 78642389 | E070 | 26732 |
| chr14 | 78642481 | 78642554 | E070 | 26888 |
| chr14 | 78644175 | 78644711 | E070 | 28582 |
| chr14 | 78644827 | 78644918 | E070 | 29234 |
| chr14 | 78647694 | 78647744 | E070 | 32101 |
| chr14 | 78647806 | 78647856 | E070 | 32213 |
| chr14 | 78650204 | 78650254 | E070 | 34611 |
| chr14 | 78650381 | 78650441 | E070 | 34788 |
| chr14 | 78650571 | 78650847 | E070 | 34978 |
| chr14 | 78651160 | 78651210 | E070 | 35567 |
| chr14 | 78660565 | 78660615 | E070 | 44972 |
| chr14 | 78661131 | 78661285 | E070 | 45538 |
| chr14 | 78661487 | 78661541 | E070 | 45894 |
| chr14 | 78662310 | 78662364 | E070 | 46717 |
| chr14 | 78662487 | 78662614 | E070 | 46894 |
| chr14 | 78662793 | 78662899 | E070 | 47200 |
| chr14 | 78663803 | 78663909 | E070 | 48210 |
| chr14 | 78664641 | 78664912 | E070 | 49048 |
| chr14 | 78598089 | 78598345 | E071 | -17248 |
| chr14 | 78598550 | 78598647 | E071 | -16946 |
| chr14 | 78618594 | 78619202 | E071 | 3001 |
| chr14 | 78623948 | 78623998 | E071 | 8355 |
| chr14 | 78624022 | 78624090 | E071 | 8429 |
| chr14 | 78624329 | 78624389 | E071 | 8736 |
| chr14 | 78597716 | 78597815 | E072 | -17778 |
| chr14 | 78598089 | 78598345 | E072 | -17248 |
| chr14 | 78636028 | 78636078 | E072 | 20435 |
| chr14 | 78636126 | 78636179 | E072 | 20533 |
| chr14 | 78644827 | 78644918 | E072 | 29234 |
| chr14 | 78598089 | 78598345 | E073 | -17248 |
| chr14 | 78598550 | 78598647 | E073 | -16946 |
| chr14 | 78655794 | 78655844 | E073 | 40201 |
| chr14 | 78597716 | 78597815 | E074 | -17778 |
| chr14 | 78598089 | 78598345 | E074 | -17248 |
| chr14 | 78598550 | 78598647 | E074 | -16946 |
| chr14 | 78588460 | 78588514 | E081 | -27079 |
| chr14 | 78588944 | 78589309 | E081 | -26284 |
| chr14 | 78589357 | 78589425 | E081 | -26168 |
| chr14 | 78589551 | 78589601 | E081 | -25992 |
| chr14 | 78589687 | 78589764 | E081 | -25829 |
| chr14 | 78647806 | 78647856 | E081 | 32213 |
| chr14 | 78662487 | 78662614 | E082 | 46894 |
| chr14 | 78662793 | 78662899 | E082 | 47200 |
Promoter Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance(-/+:Up/Downstream) |
|---|---|---|---|---|
| chr14 | 78636379 | 78637216 | E067 | 20786 |
| chr14 | 78637653 | 78637712 | E067 | 22060 |
| chr14 | 78637776 | 78637853 | E067 | 22183 |
| chr14 | 78637957 | 78638049 | E067 | 22364 |
| chr14 | 78638513 | 78638607 | E067 | 22920 |
| chr14 | 78638885 | 78638994 | E067 | 23292 |
| chr14 | 78639112 | 78639185 | E067 | 23519 |
| chr14 | 78639282 | 78639342 | E067 | 23689 |
| chr14 | 78636379 | 78637216 | E068 | 20786 |
| chr14 | 78637653 | 78637712 | E068 | 22060 |
| chr14 | 78637776 | 78637853 | E068 | 22183 |
| chr14 | 78637957 | 78638049 | E068 | 22364 |
| chr14 | 78638513 | 78638607 | E068 | 22920 |
| chr14 | 78638885 | 78638994 | E068 | 23292 |
| chr14 | 78639112 | 78639185 | E068 | 23519 |
| chr14 | 78639282 | 78639342 | E068 | 23689 |
| chr14 | 78639646 | 78639987 | E068 | 24053 |
| chr14 | 78640080 | 78640230 | E068 | 24487 |
| chr14 | 78640291 | 78640341 | E068 | 24698 |
| chr14 | 78640586 | 78640697 | E068 | 24993 |
| chr14 | 78637653 | 78637712 | E069 | 22060 |
| chr14 | 78637776 | 78637853 | E069 | 22183 |
| chr14 | 78637957 | 78638049 | E069 | 22364 |
| chr14 | 78638513 | 78638607 | E069 | 22920 |
| chr14 | 78638885 | 78638994 | E069 | 23292 |
| chr14 | 78639112 | 78639185 | E069 | 23519 |
| chr14 | 78639282 | 78639342 | E069 | 23689 |
| chr14 | 78639646 | 78639987 | E069 | 24053 |
| chr14 | 78636379 | 78637216 | E070 | 20786 |
| chr14 | 78637653 | 78637712 | E070 | 22060 |
| chr14 | 78637776 | 78637853 | E070 | 22183 |
| chr14 | 78637957 | 78638049 | E070 | 22364 |
| chr14 | 78638513 | 78638607 | E070 | 22920 |
| chr14 | 78638885 | 78638994 | E070 | 23292 |
| chr14 | 78639112 | 78639185 | E070 | 23519 |
| chr14 | 78639282 | 78639342 | E070 | 23689 |
| chr14 | 78639646 | 78639987 | E070 | 24053 |
| chr14 | 78640080 | 78640230 | E070 | 24487 |
| chr14 | 78640291 | 78640341 | E070 | 24698 |
| chr14 | 78640586 | 78640697 | E070 | 24993 |
| chr14 | 78641097 | 78641177 | E070 | 25504 |
| chr14 | 78641737 | 78641828 | E070 | 26144 |
| chr14 | 78642165 | 78642229 | E070 | 26572 |
| chr14 | 78636379 | 78637216 | E071 | 20786 |
| chr14 | 78637653 | 78637712 | E071 | 22060 |
| chr14 | 78637776 | 78637853 | E071 | 22183 |
| chr14 | 78637957 | 78638049 | E071 | 22364 |
| chr14 | 78638513 | 78638607 | E071 | 22920 |
| chr14 | 78636379 | 78637216 | E072 | 20786 |
| chr14 | 78637653 | 78637712 | E072 | 22060 |
| chr14 | 78637776 | 78637853 | E072 | 22183 |
| chr14 | 78637957 | 78638049 | E072 | 22364 |
| chr14 | 78638513 | 78638607 | E072 | 22920 |
| chr14 | 78638885 | 78638994 | E072 | 23292 |
| chr14 | 78639112 | 78639185 | E072 | 23519 |
| chr14 | 78639282 | 78639342 | E072 | 23689 |
| chr14 | 78639646 | 78639987 | E072 | 24053 |
| chr14 | 78640080 | 78640230 | E072 | 24487 |
| chr14 | 78640291 | 78640341 | E072 | 24698 |
| chr14 | 78640586 | 78640697 | E072 | 24993 |
| chr14 | 78641097 | 78641177 | E072 | 25504 |
| chr14 | 78636379 | 78637216 | E073 | 20786 |
| chr14 | 78637653 | 78637712 | E073 | 22060 |
| chr14 | 78637776 | 78637853 | E073 | 22183 |
| chr14 | 78637957 | 78638049 | E073 | 22364 |
| chr14 | 78638513 | 78638607 | E073 | 22920 |
| chr14 | 78638885 | 78638994 | E073 | 23292 |
| chr14 | 78639112 | 78639185 | E073 | 23519 |
| chr14 | 78639282 | 78639342 | E073 | 23689 |
| chr14 | 78637653 | 78637712 | E081 | 22060 |
| chr14 | 78637776 | 78637853 | E081 | 22183 |
| chr14 | 78637957 | 78638049 | E081 | 22364 |
| chr14 | 78638513 | 78638607 | E081 | 22920 |
| chr14 | 78638885 | 78638994 | E081 | 23292 |
| chr14 | 78639112 | 78639185 | E081 | 23519 |
| chr14 | 78639282 | 78639342 | E081 | 23689 |
| chr14 | 78639646 | 78639987 | E081 | 24053 |
| chr14 | 78640080 | 78640230 | E081 | 24487 |
| chr14 | 78640291 | 78640341 | E081 | 24698 |
| chr14 | 78640586 | 78640697 | E081 | 24993 |
| chr14 | 78636379 | 78637216 | E082 | 20786 |
| chr14 | 78637653 | 78637712 | E082 | 22060 |
| chr14 | 78637776 | 78637853 | E082 | 22183 |
| chr14 | 78637957 | 78638049 | E082 | 22364 |
| chr14 | 78638513 | 78638607 | E082 | 22920 |
| chr14 | 78638885 | 78638994 | E082 | 23292 |
| chr14 | 78639112 | 78639185 | E082 | 23519 |
| chr14 | 78639282 | 78639342 | E082 | 23689 |
| chr14 | 78639646 | 78639987 | E082 | 24053 |
| chr14 | 78640080 | 78640230 | E082 | 24487 |
| chr14 | 78640291 | 78640341 | E082 | 24698 |
| chr14 | 78640586 | 78640697 | E082 | 24993 |
| chr14 | 78641097 | 78641177 | E082 | 25504 |
| chr14 | 78641737 | 78641828 | E082 | 26144 |