rs4920422
Homo sapiens
C>A
LOC101927876 : Intron Variant
SNV (Single Nucleotide Variation)
A=0159 (4792/29960,GnomAD)
A=0167 (4864/29118,TOPMED)
A=0167 (836/5008,1000G)
A=0169 (652/3854,ALSPAC)
A=0156 (580/3708,TWINSUK)
A=0167 (4864/29118,TOPMED)
A=0167 (836/5008,1000G)
A=0169 (652/3854,ALSPAC)
A=0156 (580/3708,TWINSUK)
chr1:18066258 (GRCh38.p7) (1p36.13)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 1 | NC_000001.11:g.18066258C>A |
| GRCh37.p13 chr 1 | NC_000001.10:g.18392752C>A |
Gene: LOC101927876, uncharacterized LOC101927876(plus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| LINC01654 transcript | NR_125946.1:n. | N/A | Intron Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | C=0.782 | A=0.218 |
| 1000Genomes | American | Sub | 694 | C=0.870 | A=0.130 |
| 1000Genomes | East Asian | Sub | 1008 | C=0.886 | A=0.114 |
| 1000Genomes | Europe | Sub | 1006 | C=0.843 | A=0.157 |
| 1000Genomes | Global | Study-wide | 5008 | C=0.833 | A=0.167 |
| 1000Genomes | South Asian | Sub | 978 | C=0.810 | A=0.190 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | C=0.831 | A=0.169 |
| The Genome Aggregation Database | African | Sub | 8710 | C=0.795 | A=0.205 |
| The Genome Aggregation Database | American | Sub | 838 | C=0.860 | A=0.140 |
| The Genome Aggregation Database | East Asian | Sub | 1618 | C=0.912 | A=0.088 |
| The Genome Aggregation Database | Europe | Sub | 18492 | C=0.853 | A=0.146 |
| The Genome Aggregation Database | Global | Study-wide | 29960 | C=0.840 | A=0.159 |
| The Genome Aggregation Database | Other | Sub | 302 | C=0.890 | A=0.110 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | C=0.833 | A=0.167 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | C=0.844 | A=0.156 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 21314694 | Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. | Kendler KS | Alcohol Clin Exp Res |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs4920422 | 0.000252 | alcohol dependence | 21314694 |
eQTL of rs4920422 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs4920422 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
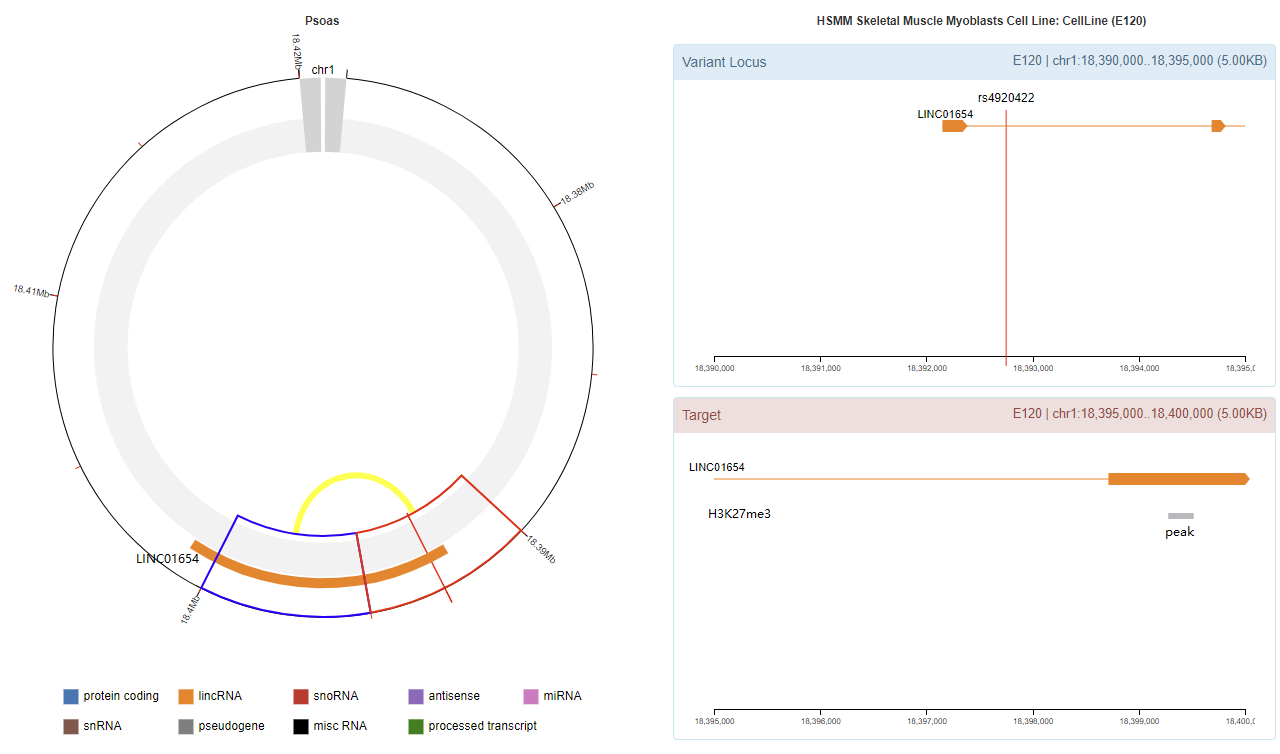
Genomic View
Chromatin Interaction
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr1 | 18385369 | 18385490 | E067 | -7262 |
| chr1 | 18385582 | 18385731 | E067 | -7021 |
| chr1 | 18385861 | 18385970 | E067 | -6782 |
| chr1 | 18427000 | 18427455 | E067 | 34248 |
| chr1 | 18438337 | 18438472 | E067 | 45585 |
| chr1 | 18438572 | 18438622 | E067 | 45820 |
| chr1 | 18385582 | 18385731 | E069 | -7021 |
| chr1 | 18438337 | 18438472 | E069 | 45585 |
| chr1 | 18438572 | 18438622 | E069 | 45820 |
| chr1 | 18358483 | 18358537 | E070 | -34215 |
| chr1 | 18358669 | 18358725 | E070 | -34027 |
| chr1 | 18384194 | 18384340 | E070 | -8412 |
| chr1 | 18384802 | 18384936 | E070 | -7816 |
| chr1 | 18385118 | 18385207 | E070 | -7545 |
| chr1 | 18385369 | 18385490 | E070 | -7262 |
| chr1 | 18386905 | 18387632 | E070 | -5120 |
| chr1 | 18390724 | 18390774 | E070 | -1978 |
| chr1 | 18391062 | 18391321 | E070 | -1431 |
| chr1 | 18398553 | 18398611 | E070 | 5801 |
| chr1 | 18398633 | 18398805 | E070 | 5881 |
| chr1 | 18404954 | 18405028 | E070 | 12202 |
| chr1 | 18405225 | 18405335 | E070 | 12473 |
| chr1 | 18406450 | 18406528 | E070 | 13698 |
| chr1 | 18406647 | 18406754 | E070 | 13895 |
| chr1 | 18407118 | 18407293 | E070 | 14366 |
| chr1 | 18407387 | 18407433 | E070 | 14635 |
| chr1 | 18417152 | 18417620 | E070 | 24400 |
| chr1 | 18417854 | 18417917 | E070 | 25102 |
| chr1 | 18417925 | 18417975 | E070 | 25173 |
| chr1 | 18418142 | 18418271 | E070 | 25390 |
| chr1 | 18438337 | 18438472 | E070 | 45585 |
| chr1 | 18385582 | 18385731 | E071 | -7021 |
| chr1 | 18438337 | 18438472 | E071 | 45585 |
| chr1 | 18438572 | 18438622 | E071 | 45820 |
| chr1 | 18433391 | 18433507 | E072 | 40639 |
| chr1 | 18438337 | 18438472 | E072 | 45585 |
| chr1 | 18438572 | 18438622 | E072 | 45820 |
| chr1 | 18385118 | 18385207 | E073 | -7545 |
| chr1 | 18385369 | 18385490 | E073 | -7262 |
| chr1 | 18385582 | 18385731 | E073 | -7021 |
| chr1 | 18438337 | 18438472 | E073 | 45585 |
| chr1 | 18438572 | 18438622 | E073 | 45820 |
| chr1 | 18378954 | 18379517 | E081 | -13235 |
| chr1 | 18379583 | 18379778 | E081 | -12974 |
| chr1 | 18384802 | 18384936 | E081 | -7816 |
| chr1 | 18385118 | 18385207 | E081 | -7545 |
| chr1 | 18385369 | 18385490 | E081 | -7262 |
| chr1 | 18385582 | 18385731 | E081 | -7021 |
| chr1 | 18386905 | 18387632 | E081 | -5120 |
| chr1 | 18398553 | 18398611 | E081 | 5801 |
| chr1 | 18398633 | 18398805 | E081 | 5881 |
| chr1 | 18399702 | 18399788 | E081 | 6950 |
| chr1 | 18400338 | 18400769 | E081 | 7586 |
| chr1 | 18400801 | 18400942 | E081 | 8049 |
| chr1 | 18405225 | 18405335 | E081 | 12473 |
| chr1 | 18406450 | 18406528 | E081 | 13698 |
| chr1 | 18406647 | 18406754 | E081 | 13895 |
| chr1 | 18407118 | 18407293 | E081 | 14366 |
| chr1 | 18407387 | 18407433 | E081 | 14635 |
| chr1 | 18417152 | 18417620 | E081 | 24400 |
| chr1 | 18417854 | 18417917 | E081 | 25102 |
| chr1 | 18417925 | 18417975 | E081 | 25173 |
| chr1 | 18418142 | 18418271 | E081 | 25390 |
| chr1 | 18418303 | 18418573 | E081 | 25551 |
| chr1 | 18425555 | 18425864 | E081 | 32803 |
| chr1 | 18426357 | 18426425 | E081 | 33605 |
| chr1 | 18433391 | 18433507 | E081 | 40639 |
| chr1 | 18433593 | 18433730 | E081 | 40841 |
| chr1 | 18438337 | 18438472 | E081 | 45585 |
| chr1 | 18438572 | 18438622 | E081 | 45820 |
| chr1 | 18386905 | 18387632 | E082 | -5120 |
| chr1 | 18398051 | 18398101 | E082 | 5299 |
| chr1 | 18398553 | 18398611 | E082 | 5801 |
| chr1 | 18398633 | 18398805 | E082 | 5881 |
| chr1 | 18406450 | 18406528 | E082 | 13698 |
| chr1 | 18406647 | 18406754 | E082 | 13895 |
| chr1 | 18417152 | 18417620 | E082 | 24400 |
| chr1 | 18417854 | 18417917 | E082 | 25102 |
| chr1 | 18417925 | 18417975 | E082 | 25173 |
| chr1 | 18418142 | 18418271 | E082 | 25390 |
| chr1 | 18418303 | 18418573 | E082 | 25551 |
Promoter Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance(-/+:Up/Downstream) |
|---|---|---|---|---|
| chr1 | 18433793 | 18434371 | E067 | 41041 |
| chr1 | 18434620 | 18435196 | E067 | 41868 |
| chr1 | 18435251 | 18435388 | E067 | 42499 |
| chr1 | 18435458 | 18435574 | E067 | 42706 |
| chr1 | 18435659 | 18435823 | E067 | 42907 |
| chr1 | 18436479 | 18436531 | E067 | 43727 |
| chr1 | 18436546 | 18437025 | E067 | 43794 |
| chr1 | 18437039 | 18437348 | E067 | 44287 |
| chr1 | 18437562 | 18437862 | E067 | 44810 |
| chr1 | 18433793 | 18434371 | E068 | 41041 |
| chr1 | 18435251 | 18435388 | E068 | 42499 |
| chr1 | 18435458 | 18435574 | E068 | 42706 |
| chr1 | 18436479 | 18436531 | E068 | 43727 |
| chr1 | 18436546 | 18437025 | E068 | 43794 |
| chr1 | 18437039 | 18437348 | E068 | 44287 |
| chr1 | 18437562 | 18437862 | E068 | 44810 |
| chr1 | 18433793 | 18434371 | E069 | 41041 |
| chr1 | 18434620 | 18435196 | E069 | 41868 |
| chr1 | 18436546 | 18437025 | E069 | 43794 |
| chr1 | 18437039 | 18437348 | E069 | 44287 |
| chr1 | 18437562 | 18437862 | E069 | 44810 |
| chr1 | 18433793 | 18434371 | E070 | 41041 |
| chr1 | 18434620 | 18435196 | E071 | 41868 |
| chr1 | 18434620 | 18435196 | E072 | 41868 |
| chr1 | 18435251 | 18435388 | E072 | 42499 |
| chr1 | 18435458 | 18435574 | E072 | 42706 |
| chr1 | 18435659 | 18435823 | E072 | 42907 |
| chr1 | 18436479 | 18436531 | E072 | 43727 |
| chr1 | 18436546 | 18437025 | E072 | 43794 |
| chr1 | 18437039 | 18437348 | E072 | 44287 |
| chr1 | 18437562 | 18437862 | E072 | 44810 |
| chr1 | 18433793 | 18434371 | E073 | 41041 |
| chr1 | 18434620 | 18435196 | E073 | 41868 |
| chr1 | 18435251 | 18435388 | E073 | 42499 |
| chr1 | 18435458 | 18435574 | E073 | 42706 |
| chr1 | 18435659 | 18435823 | E073 | 42907 |
| chr1 | 18436479 | 18436531 | E073 | 43727 |
| chr1 | 18436546 | 18437025 | E073 | 43794 |
| chr1 | 18437039 | 18437348 | E073 | 44287 |
| chr1 | 18433793 | 18434371 | E074 | 41041 |
| chr1 | 18436479 | 18436531 | E074 | 43727 |
| chr1 | 18436546 | 18437025 | E074 | 43794 |
| chr1 | 18437039 | 18437348 | E074 | 44287 |
| chr1 | 18433793 | 18434371 | E082 | 41041 |
| chr1 | 18434620 | 18435196 | E082 | 41868 |
| chr1 | 18436479 | 18436531 | E082 | 43727 |
| chr1 | 18437562 | 18437862 | E082 | 44810 |