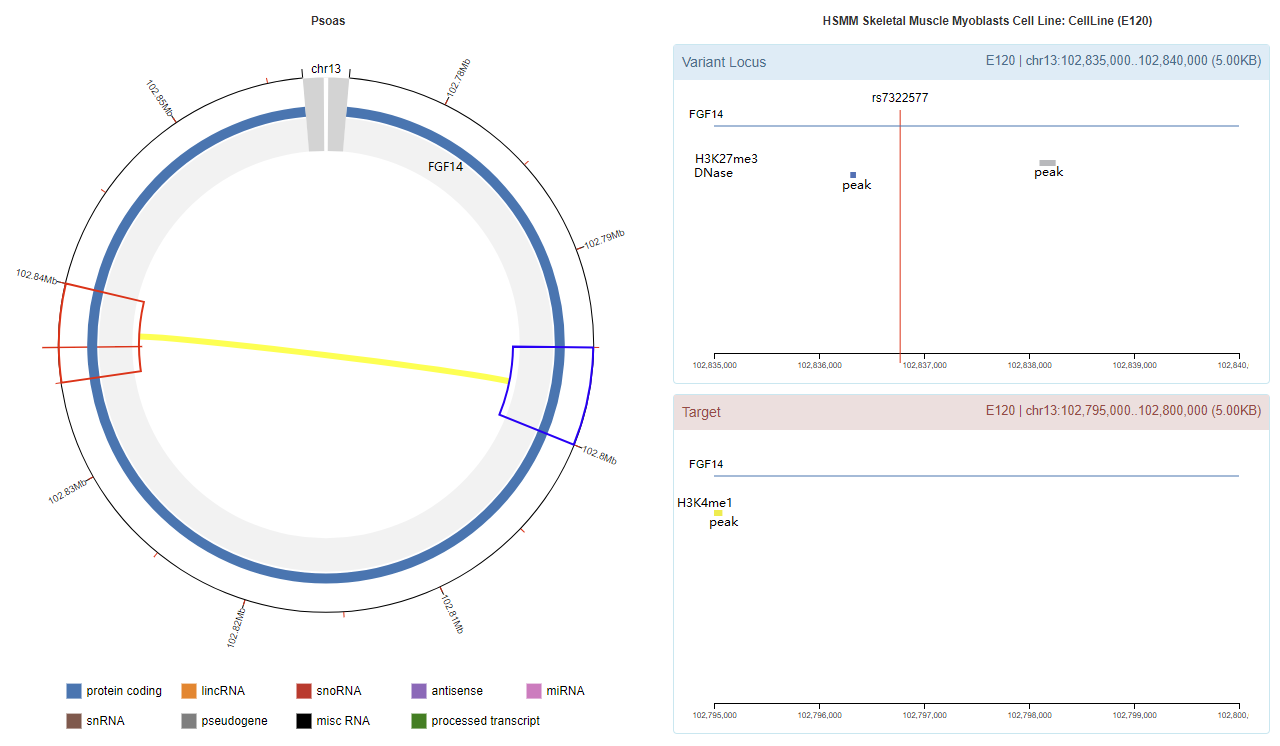
rs7322577
Homo sapiens
T>A / T>G
FGF14 : Intron Variant
SNV (Single Nucleotide Variation)
G=0191 (5733/29918,GnomAD)
G=0185 (5396/29118,TOPMED)
G=0194 (971/5008,1000G)
G=0185 (5396/29118,TOPMED)
G=0194 (971/5008,1000G)
chr13:102184424 (GRCh38.p7) (13q33.1)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 13 | NC_000013.11:g.102184424T>A |
| GRCh38.p7 chr 13 | NC_000013.11:g.102184424T>G |
| GRCh37.p13 chr 13 | NC_000013.10:g.102836774T>A |
| GRCh37.p13 chr 13 | NC_000013.10:g.102836774T>G |
| FGF14 RefSeqGene | NG_008317.1:g.222351A>T |
| FGF14 RefSeqGene | NG_008317.1:g.222351A>C |
Gene: FGF14, fibroblast growth factor 14(minus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| FGF14 transcript variant 3 | NM_001321931.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 4 | NM_001321932.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 5 | NM_001321933.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 6 | NM_001321934.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 7 | NM_001321935.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 8 | NM_001321936.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 9 | NM_001321937.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 10 | NM_001321938.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 11 | NM_001321939.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 12 | NM_001321940.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 13 | NM_001321941.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 14 | NM_001321942.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 15 | NM_001321943.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 16 | NM_001321944.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 17 | NM_001321945.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 18 | NM_001321946.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 19 | NM_001321947.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 20 | NM_001321948.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 21 | NM_001321949.1:c. | N/A | Intron Variant |
| FGF14 transcript variant 2 | NM_175929.2:c. | N/A | Intron Variant |
| FGF14 transcript variant 1 | NM_004115.3:c. | N/A | Genic Upstream Transcript Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | T=0.840 | G=0.160 |
| 1000Genomes | American | Sub | 694 | T=0.880 | G=0.120 |
| 1000Genomes | East Asian | Sub | 1008 | T=0.773 | G=0.227 |
| 1000Genomes | Europe | Sub | 1006 | T=0.783 | G=0.217 |
| 1000Genomes | Global | Study-wide | 5008 | T=0.806 | G=0.194 |
| 1000Genomes | South Asian | Sub | 978 | T=0.760 | G=0.240 |
| The Genome Aggregation Database | African | Sub | 8708 | T=0.827 | G=0.173 |
| The Genome Aggregation Database | American | Sub | 836 | T=0.900 | G=0.100 |
| The Genome Aggregation Database | East Asian | Sub | 1594 | T=0.775 | G=0.225 |
| The Genome Aggregation Database | Europe | Sub | 18478 | T=0.797 | G=0.202 |
| The Genome Aggregation Database | Global | Study-wide | 29918 | T=0.808 | G=0.191 |
| The Genome Aggregation Database | Other | Sub | 302 | T=0.840 | G=0.160 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | T=0.814 | G=0.185 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 20201924 | Genome-wide association study of alcohol dependence implicates a region on chromosome 11. | Edenberg HJ | Alcohol Clin Exp Res |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs7322577 | 0.000419 | alcohol dependence | 20201924 |
eQTL of rs7322577 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs7322577 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
Genomic View
Chromatin Interaction