rs7779286
Homo sapiens
A>G
LOC107986765 : Intron Variant
SNV (Single Nucleotide Variation)
A==0186 (5577/29964,GnomAD)
A==0164 (4785/29118,TOPMED)
A==0080 (402/5008,1000G)
A==0248 (956/3854,ALSPAC)
A==0260 (964/3708,TWINSUK)
A==0164 (4785/29118,TOPMED)
A==0080 (402/5008,1000G)
A==0248 (956/3854,ALSPAC)
A==0260 (964/3708,TWINSUK)
chr7:7936612 (GRCh38.p7) (7p21.3)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 7 | NC_000007.14:g.7936612A>G |
| GRCh37.p13 chr 7 | NC_000007.13:g.7976243A>G |
Gene: LOC107986765, uncharacterized LOC107986765(minus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| LOC107986765 transcript variant X2 | XR_001745079.1:n. | N/A | Intron Variant |
| LOC107986765 transcript variant X1 | XR_001745078.1:n. | N/A | Genic Upstream Transcript Variant |
| LOC107986765 transcript variant X3 | XR_001745080.1:n. | N/A | Genic Upstream Transcript Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | A=0.041 | G=0.959 |
| 1000Genomes | American | Sub | 694 | A=0.110 | G=0.890 |
| 1000Genomes | East Asian | Sub | 1008 | A=0.001 | G=0.999 |
| 1000Genomes | Europe | Sub | 1006 | A=0.230 | G=0.770 |
| 1000Genomes | Global | Study-wide | 5008 | A=0.080 | G=0.920 |
| 1000Genomes | South Asian | Sub | 978 | A=0.040 | G=0.960 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | A=0.248 | G=0.752 |
| The Genome Aggregation Database | African | Sub | 8730 | A=0.064 | G=0.936 |
| The Genome Aggregation Database | American | Sub | 838 | A=0.140 | G=0.860 |
| The Genome Aggregation Database | East Asian | Sub | 1612 | A=0.000 | G=1.000 |
| The Genome Aggregation Database | Europe | Sub | 18482 | A=0.259 | G=0.740 |
| The Genome Aggregation Database | Global | Study-wide | 29964 | A=0.186 | G=0.813 |
| The Genome Aggregation Database | Other | Sub | 302 | A=0.330 | G=0.670 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | A=0.164 | G=0.835 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | A=0.260 | G=0.740 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 20201924 | Genome-wide association study of alcohol dependence implicates a region on chromosome 11. | Edenberg HJ | Alcohol Clin Exp Res |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs7779286 | 0.00078 | alcohol dependence | 20201924 |
eQTL of rs7779286 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs7779286 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
Genomic View
Chromatin Interaction
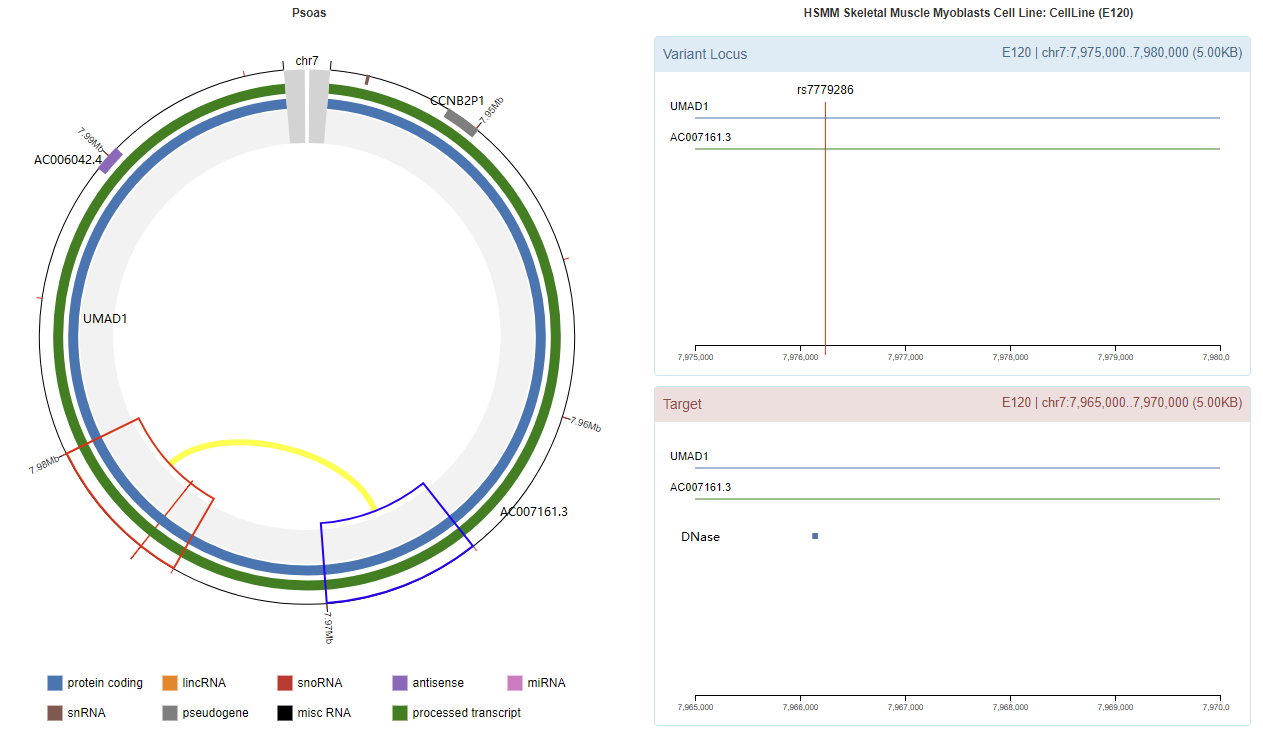
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr7 | 8014753 | 8014811 | E067 | 38510 |
| chr7 | 8014914 | 8015039 | E067 | 38671 |
| chr7 | 8017441 | 8017720 | E067 | 41198 |
| chr7 | 7959627 | 7959715 | E068 | -16528 |
| chr7 | 7959746 | 7959799 | E068 | -16444 |
| chr7 | 7961621 | 7961734 | E068 | -14509 |
| chr7 | 7961741 | 7961835 | E068 | -14408 |
| chr7 | 7961932 | 7962329 | E068 | -13914 |
| chr7 | 7962740 | 7962861 | E068 | -13382 |
| chr7 | 7963258 | 7963338 | E068 | -12905 |
| chr7 | 7963347 | 7963567 | E068 | -12676 |
| chr7 | 7983606 | 7985268 | E068 | 7363 |
| chr7 | 8012512 | 8012593 | E068 | 36269 |
| chr7 | 8012652 | 8012756 | E068 | 36409 |
| chr7 | 8014753 | 8014811 | E068 | 38510 |
| chr7 | 8014914 | 8015039 | E068 | 38671 |
| chr7 | 8015088 | 8015141 | E068 | 38845 |
| chr7 | 7932305 | 7932355 | E069 | -43888 |
| chr7 | 7961741 | 7961835 | E069 | -14408 |
| chr7 | 7961932 | 7962329 | E069 | -13914 |
| chr7 | 7962402 | 7962604 | E069 | -13639 |
| chr7 | 7962622 | 7962695 | E069 | -13548 |
| chr7 | 7962740 | 7962861 | E069 | -13382 |
| chr7 | 7963258 | 7963338 | E069 | -12905 |
| chr7 | 7963347 | 7963567 | E069 | -12676 |
| chr7 | 7934298 | 7934383 | E070 | -41860 |
| chr7 | 7934505 | 7934622 | E070 | -41621 |
| chr7 | 7934646 | 7935135 | E070 | -41108 |
| chr7 | 7935236 | 7935341 | E070 | -40902 |
| chr7 | 7935414 | 7935652 | E070 | -40591 |
| chr7 | 7959880 | 7960008 | E070 | -16235 |
| chr7 | 7960088 | 7960458 | E070 | -15785 |
| chr7 | 7960537 | 7960581 | E070 | -15662 |
| chr7 | 7960618 | 7960719 | E070 | -15524 |
| chr7 | 7960810 | 7960878 | E070 | -15365 |
| chr7 | 7961046 | 7961112 | E070 | -15131 |
| chr7 | 7961621 | 7961734 | E070 | -14509 |
| chr7 | 7961741 | 7961835 | E070 | -14408 |
| chr7 | 7961932 | 7962329 | E070 | -13914 |
| chr7 | 7962402 | 7962604 | E070 | -13639 |
| chr7 | 7962622 | 7962695 | E070 | -13548 |
| chr7 | 7962740 | 7962861 | E070 | -13382 |
| chr7 | 7963258 | 7963338 | E070 | -12905 |
| chr7 | 7963347 | 7963567 | E070 | -12676 |
| chr7 | 7963618 | 7963729 | E070 | -12514 |
| chr7 | 7963745 | 7963896 | E070 | -12347 |
| chr7 | 7963981 | 7964181 | E070 | -12062 |
| chr7 | 8017106 | 8017193 | E070 | 40863 |
| chr7 | 8017441 | 8017720 | E070 | 41198 |
| chr7 | 8019154 | 8019299 | E070 | 42911 |
| chr7 | 8019337 | 8019483 | E070 | 43094 |
| chr7 | 8019559 | 8019625 | E070 | 43316 |
| chr7 | 8019898 | 8020085 | E070 | 43655 |
| chr7 | 7934298 | 7934383 | E071 | -41860 |
| chr7 | 7961621 | 7961734 | E071 | -14509 |
| chr7 | 7961741 | 7961835 | E071 | -14408 |
| chr7 | 7961932 | 7962329 | E071 | -13914 |
| chr7 | 7963618 | 7963729 | E071 | -12514 |
| chr7 | 8012512 | 8012593 | E071 | 36269 |
| chr7 | 8012652 | 8012756 | E071 | 36409 |
| chr7 | 8012971 | 8013274 | E071 | 36728 |
| chr7 | 8013314 | 8013418 | E071 | 37071 |
| chr7 | 7932305 | 7932355 | E072 | -43888 |
| chr7 | 7960088 | 7960458 | E072 | -15785 |
| chr7 | 7960537 | 7960581 | E072 | -15662 |
| chr7 | 7960618 | 7960719 | E072 | -15524 |
| chr7 | 7960810 | 7960878 | E072 | -15365 |
| chr7 | 7961621 | 7961734 | E072 | -14509 |
| chr7 | 7961741 | 7961835 | E072 | -14408 |
| chr7 | 7961932 | 7962329 | E072 | -13914 |
| chr7 | 7962402 | 7962604 | E072 | -13639 |
| chr7 | 8017106 | 8017193 | E073 | 40863 |
| chr7 | 7959004 | 7959554 | E074 | -16689 |
| chr7 | 7959627 | 7959715 | E074 | -16528 |
| chr7 | 7959746 | 7959799 | E074 | -16444 |
| chr7 | 7959880 | 7960008 | E074 | -16235 |
| chr7 | 7960088 | 7960458 | E074 | -15785 |
| chr7 | 7960537 | 7960581 | E074 | -15662 |
| chr7 | 7960618 | 7960719 | E074 | -15524 |
| chr7 | 7960810 | 7960878 | E074 | -15365 |
| chr7 | 7961046 | 7961112 | E074 | -15131 |
| chr7 | 7961392 | 7961442 | E074 | -14801 |
| chr7 | 7961621 | 7961734 | E074 | -14509 |
| chr7 | 7961741 | 7961835 | E074 | -14408 |
| chr7 | 7961932 | 7962329 | E074 | -13914 |
| chr7 | 7963618 | 7963729 | E074 | -12514 |
| chr7 | 8012512 | 8012593 | E074 | 36269 |
| chr7 | 8012971 | 8013274 | E074 | 36728 |
| chr7 | 7932305 | 7932355 | E081 | -43888 |
| chr7 | 7933254 | 7933304 | E081 | -42939 |
| chr7 | 8012512 | 8012593 | E081 | 36269 |
| chr7 | 8012652 | 8012756 | E081 | 36409 |
| chr7 | 8012971 | 8013274 | E081 | 36728 |
| chr7 | 8012971 | 8013274 | E082 | 36728 |
Promoter Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance(-/+:Up/Downstream) |
|---|---|---|---|---|
| chr7 | 8007483 | 8011594 | E067 | 31240 |
| chr7 | 8007483 | 8011594 | E068 | 31240 |
| chr7 | 8007483 | 8011594 | E069 | 31240 |
| chr7 | 8007483 | 8011594 | E070 | 31240 |
| chr7 | 8007326 | 8007470 | E071 | 31083 |
| chr7 | 8007483 | 8011594 | E071 | 31240 |
| chr7 | 8007483 | 8011594 | E072 | 31240 |
| chr7 | 8007483 | 8011594 | E073 | 31240 |
| chr7 | 8007483 | 8011594 | E074 | 31240 |
| chr7 | 8007483 | 8011594 | E081 | 31240 |
| chr7 | 8007483 | 8011594 | E082 | 31240 |