rs8177178
Homo sapiens
G>A
TF : Intron Variant
SNV (Single Nucleotide Variation)
A=0332 (9952/29922,GnomAD)
A=0333 (9706/29118,TOPMED)
A=0370 (1853/5008,1000G)
A=0320 (1232/3854,ALSPAC)
A=0305 (1132/3708,TWINSUK)
A=0333 (9706/29118,TOPMED)
A=0370 (1853/5008,1000G)
A=0320 (1232/3854,ALSPAC)
A=0305 (1132/3708,TWINSUK)
chr3:133744428 (GRCh38.p7) (3q22.1)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 3 | NC_000003.12:g.133744428G>A |
| GRCh37.p13 chr 3 | NC_000003.11:g.133463272G>A |
| TF RefSeqGene | NG_013080.1:g.3296G>A |
Gene: TF, transferrin(plus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| TF transcript variant 1 | NM_001063.3:c. | N/A | Genic Upstream Transcript Variant |
| TF transcript variant X1 | XM_017007089.1:c. | N/A | Intron Variant |
| TF transcript variant X2 | XM_017007090.1:c. | N/A | Intron Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | G=0.679 | A=0.321 |
| 1000Genomes | American | Sub | 694 | G=0.600 | A=0.400 |
| 1000Genomes | East Asian | Sub | 1008 | G=0.579 | A=0.421 |
| 1000Genomes | Europe | Sub | 1006 | G=0.679 | A=0.321 |
| 1000Genomes | Global | Study-wide | 5008 | G=0.630 | A=0.370 |
| 1000Genomes | South Asian | Sub | 978 | G=0.590 | A=0.410 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | G=0.680 | A=0.320 |
| The Genome Aggregation Database | African | Sub | 8706 | G=0.648 | A=0.352 |
| The Genome Aggregation Database | American | Sub | 838 | G=0.550 | A=0.450 |
| The Genome Aggregation Database | East Asian | Sub | 1618 | G=0.611 | A=0.389 |
| The Genome Aggregation Database | Europe | Sub | 18458 | G=0.685 | A=0.314 |
| The Genome Aggregation Database | Global | Study-wide | 29922 | G=0.667 | A=0.332 |
| The Genome Aggregation Database | Other | Sub | 302 | G=0.780 | A=0.220 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | G=0.666 | A=0.333 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | G=0.695 | A=0.305 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 19673882 | A novel association between a SNP in CYBRD1 and serum ferritin levels in a cohort study of HFE hereditary haemochromatosis. | Constantine CC | Br J Haematol |
| 21665994 | Genome-wide association study identifies two loci strongly affecting transferrin glycosylation. | Kutalik Z | Hum Mol Genet |
| 23089144 | An association of transferrin gene polymorphism and serum transferrin levels with age-related macular degeneration. | Wysokinski D | Exp Eye Res |
| 25097799 | Developments in Ocular Genetics: 2013 Annual Review. | Aboobakar IF | Asia Pac J Ophthalmol (Phila) |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs8177178 | 4.02E-12 | alcohol consumption | 21665994 |
eQTL of rs8177178 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs8177178 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value |
|---|---|---|---|---|
| cg08048268 | chr3:133502702 | -0.120490848109944 | 6.9020e-18 | |
| cg01448562 | chr3:133502909 | -0.0527264162818601 | 1.4158e-17 | |
| cg16414030 | chr3:133502952 | -0.0776625247990655 | 9.8827e-16 | |
| cg16275903 | chr3:133524006 | SRPRB | 0.0500724108221123 | 7.5914e-15 |
| cg08439880 | chr3:133502540 | -0.0607451338499631 | 1.9325e-12 | |
| cg20276088 | chr3:133502917 | -0.0300747717526692 | 3.5207e-12 | |
| cg11941060 | chr3:133502564 | -0.0523216624930985 | 7.4788e-11 |
Genomic View
Chromatin Interaction
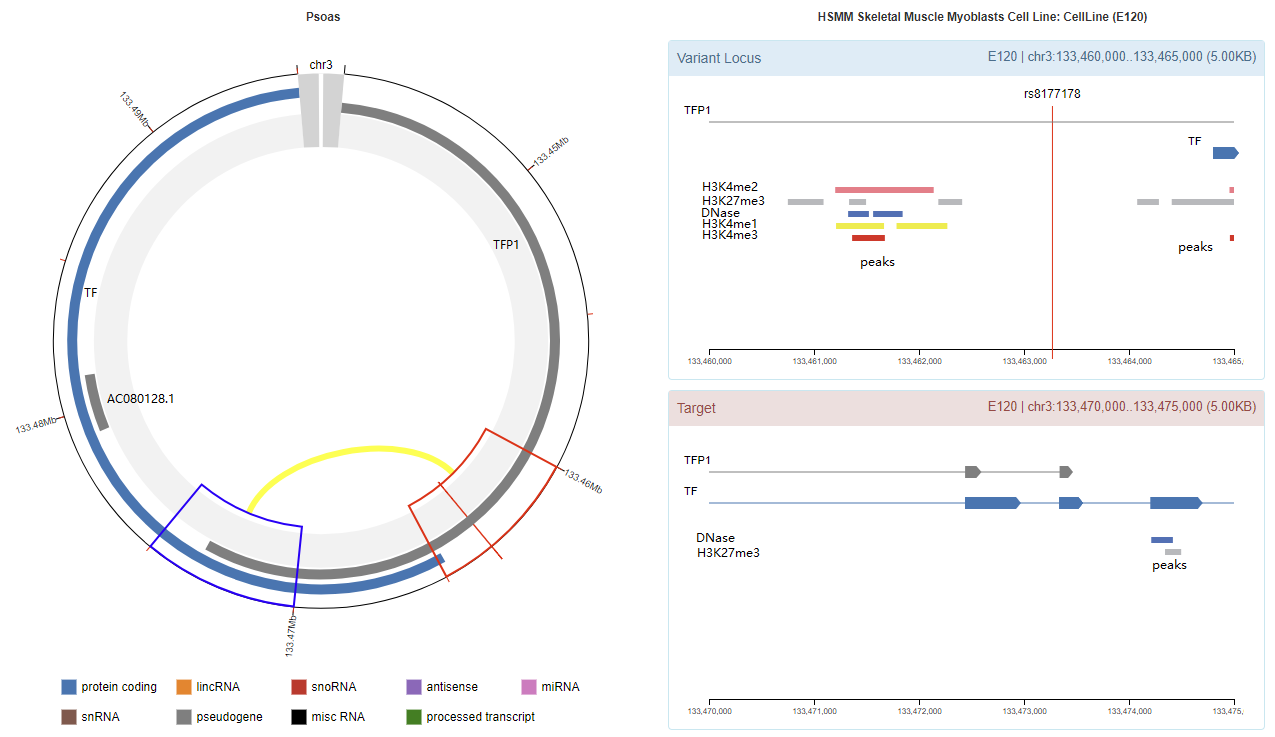
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr3 | 133431016 | 133431089 | E067 | -32183 |
| chr3 | 133436424 | 133436504 | E067 | -26768 |
| chr3 | 133461397 | 133461916 | E067 | -1356 |
| chr3 | 133461945 | 133462055 | E067 | -1217 |
| chr3 | 133464069 | 133464119 | E067 | 797 |
| chr3 | 133464448 | 133464526 | E067 | 1176 |
| chr3 | 133482923 | 133483028 | E067 | 19651 |
| chr3 | 133483054 | 133483594 | E067 | 19782 |
| chr3 | 133483998 | 133484070 | E067 | 20726 |
| chr3 | 133436424 | 133436504 | E068 | -26768 |
| chr3 | 133464069 | 133464119 | E068 | 797 |
| chr3 | 133482562 | 133482616 | E068 | 19290 |
| chr3 | 133482923 | 133483028 | E068 | 19651 |
| chr3 | 133483054 | 133483594 | E068 | 19782 |
| chr3 | 133431016 | 133431089 | E069 | -32183 |
| chr3 | 133436424 | 133436504 | E069 | -26768 |
| chr3 | 133461397 | 133461916 | E069 | -1356 |
| chr3 | 133461945 | 133462055 | E069 | -1217 |
| chr3 | 133464069 | 133464119 | E069 | 797 |
| chr3 | 133473014 | 133473073 | E069 | 9742 |
| chr3 | 133473315 | 133473659 | E069 | 10043 |
| chr3 | 133476260 | 133476458 | E069 | 12988 |
| chr3 | 133482562 | 133482616 | E069 | 19290 |
| chr3 | 133482923 | 133483028 | E069 | 19651 |
| chr3 | 133483054 | 133483594 | E069 | 19782 |
| chr3 | 133483998 | 133484070 | E069 | 20726 |
| chr3 | 133484337 | 133484387 | E069 | 21065 |
| chr3 | 133482923 | 133483028 | E070 | 19651 |
| chr3 | 133483054 | 133483594 | E070 | 19782 |
| chr3 | 133431016 | 133431089 | E071 | -32183 |
| chr3 | 133436424 | 133436504 | E071 | -26768 |
| chr3 | 133461397 | 133461916 | E071 | -1356 |
| chr3 | 133461945 | 133462055 | E071 | -1217 |
| chr3 | 133464069 | 133464119 | E071 | 797 |
| chr3 | 133473014 | 133473073 | E071 | 9742 |
| chr3 | 133473315 | 133473659 | E071 | 10043 |
| chr3 | 133482562 | 133482616 | E071 | 19290 |
| chr3 | 133482923 | 133483028 | E071 | 19651 |
| chr3 | 133483054 | 133483594 | E071 | 19782 |
| chr3 | 133483998 | 133484070 | E071 | 20726 |
| chr3 | 133484337 | 133484387 | E071 | 21065 |
| chr3 | 133431016 | 133431089 | E072 | -32183 |
| chr3 | 133461397 | 133461916 | E072 | -1356 |
| chr3 | 133461945 | 133462055 | E072 | -1217 |
| chr3 | 133464069 | 133464119 | E072 | 797 |
| chr3 | 133464448 | 133464526 | E072 | 1176 |
| chr3 | 133473014 | 133473073 | E072 | 9742 |
| chr3 | 133482923 | 133483028 | E072 | 19651 |
| chr3 | 133483054 | 133483594 | E072 | 19782 |
| chr3 | 133483998 | 133484070 | E072 | 20726 |
| chr3 | 133484337 | 133484387 | E072 | 21065 |
| chr3 | 133436424 | 133436504 | E073 | -26768 |
| chr3 | 133461397 | 133461916 | E073 | -1356 |
| chr3 | 133461945 | 133462055 | E073 | -1217 |
| chr3 | 133464448 | 133464526 | E073 | 1176 |
| chr3 | 133482923 | 133483028 | E073 | 19651 |
| chr3 | 133483054 | 133483594 | E073 | 19782 |
| chr3 | 133431016 | 133431089 | E074 | -32183 |
| chr3 | 133436424 | 133436504 | E074 | -26768 |
| chr3 | 133461397 | 133461916 | E074 | -1356 |
| chr3 | 133461945 | 133462055 | E074 | -1217 |
| chr3 | 133464069 | 133464119 | E074 | 797 |
| chr3 | 133473014 | 133473073 | E074 | 9742 |
| chr3 | 133473315 | 133473659 | E074 | 10043 |
| chr3 | 133476260 | 133476458 | E074 | 12988 |
| chr3 | 133482562 | 133482616 | E074 | 19290 |
| chr3 | 133482923 | 133483028 | E074 | 19651 |
| chr3 | 133483054 | 133483594 | E074 | 19782 |
| chr3 | 133483998 | 133484070 | E074 | 20726 |
| chr3 | 133484337 | 133484387 | E074 | 21065 |
| chr3 | 133464448 | 133464526 | E082 | 1176 |
Promoter Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance(-/+:Up/Downstream) |
|---|---|---|---|---|
| chr3 | 133464975 | 133465152 | E067 | 1703 |
| chr3 | 133465195 | 133465439 | E067 | 1923 |
| chr3 | 133465691 | 133465761 | E067 | 2419 |
| chr3 | 133468272 | 133468322 | E067 | 5000 |
| chr3 | 133464975 | 133465152 | E068 | 1703 |
| chr3 | 133465195 | 133465439 | E068 | 1923 |
| chr3 | 133465691 | 133465761 | E068 | 2419 |
| chr3 | 133468272 | 133468322 | E068 | 5000 |
| chr3 | 133464975 | 133465152 | E069 | 1703 |
| chr3 | 133465195 | 133465439 | E069 | 1923 |
| chr3 | 133465691 | 133465761 | E069 | 2419 |
| chr3 | 133468272 | 133468322 | E069 | 5000 |
| chr3 | 133465195 | 133465439 | E070 | 1923 |
| chr3 | 133464975 | 133465152 | E071 | 1703 |
| chr3 | 133465195 | 133465439 | E071 | 1923 |
| chr3 | 133465691 | 133465761 | E071 | 2419 |
| chr3 | 133468272 | 133468322 | E071 | 5000 |
| chr3 | 133464975 | 133465152 | E072 | 1703 |
| chr3 | 133465195 | 133465439 | E072 | 1923 |
| chr3 | 133465691 | 133465761 | E072 | 2419 |
| chr3 | 133468272 | 133468322 | E072 | 5000 |
| chr3 | 133464975 | 133465152 | E073 | 1703 |
| chr3 | 133465195 | 133465439 | E073 | 1923 |
| chr3 | 133465691 | 133465761 | E073 | 2419 |
| chr3 | 133468272 | 133468322 | E073 | 5000 |
| chr3 | 133464975 | 133465152 | E074 | 1703 |
| chr3 | 133465195 | 133465439 | E074 | 1923 |
| chr3 | 133465691 | 133465761 | E074 | 2419 |
| chr3 | 133468272 | 133468322 | E074 | 5000 |
| chr3 | 133464975 | 133465152 | E081 | 1703 |
| chr3 | 133464975 | 133465152 | E082 | 1703 |
| chr3 | 133465195 | 133465439 | E082 | 1923 |