rs8177213
Homo sapiens
A>C
TF : Intron Variant
SNV (Single Nucleotide Variation)
C=0240 (7178/29908,GnomAD)
C=0241 (7032/29118,TOPMED)
C=0216 (1082/5008,1000G)
C=0287 (1105/3854,ALSPAC)
C=0289 (1072/3708,TWINSUK)
C=0241 (7032/29118,TOPMED)
C=0216 (1082/5008,1000G)
C=0287 (1105/3854,ALSPAC)
C=0289 (1072/3708,TWINSUK)
chr3:133753383 (GRCh38.p7) (3q22.1)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 3 | NC_000003.12:g.133753383A>C |
| GRCh37.p13 chr 3 | NC_000003.11:g.133472227A>C |
| TF RefSeqGene | NG_013080.1:g.12251A>C |
Gene: TF, transferrin(plus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| TF transcript variant 1 | NM_001063.3:c. | N/A | Intron Variant |
| TF transcript variant X1 | XM_017007089.1:c. | N/A | Intron Variant |
| TF transcript variant X2 | XM_017007090.1:c. | N/A | Intron Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | A=0.792 | C=0.208 |
| 1000Genomes | American | Sub | 694 | A=0.760 | C=0.240 |
| 1000Genomes | East Asian | Sub | 1008 | A=0.760 | C=0.240 |
| 1000Genomes | Europe | Sub | 1006 | A=0.759 | C=0.241 |
| 1000Genomes | Global | Study-wide | 5008 | A=0.784 | C=0.216 |
| 1000Genomes | South Asian | Sub | 978 | A=0.840 | C=0.160 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | A=0.713 | C=0.287 |
| The Genome Aggregation Database | African | Sub | 8698 | A=0.785 | C=0.215 |
| The Genome Aggregation Database | American | Sub | 836 | A=0.770 | C=0.230 |
| The Genome Aggregation Database | East Asian | Sub | 1616 | A=0.780 | C=0.220 |
| The Genome Aggregation Database | Europe | Sub | 18460 | A=0.747 | C=0.252 |
| The Genome Aggregation Database | Global | Study-wide | 29908 | A=0.760 | C=0.240 |
| The Genome Aggregation Database | Other | Sub | 298 | A=0.670 | C=0.330 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | A=0.758 | C=0.241 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | A=0.711 | C=0.289 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 21665994 | Genome-wide association study identifies two loci strongly affecting transferrin glycosylation. | Kutalik Z | Hum Mol Genet |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs8177213 | 2.92E-07 | alcohol consumption | 21665994 |
eQTL of rs8177213 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs8177213 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value |
|---|---|---|---|---|
| cg08048268 | chr3:133502702 | 0.117219594326722 | 4.3708e-11 | |
| cg01448562 | chr3:133502909 | 0.0463115739718659 | 5.8377e-9 | |
| cg16275903 | chr3:133524006 | SRPRB | -0.0472046118606483 | 6.9198e-9 |
Genomic View
Chromatin Interaction
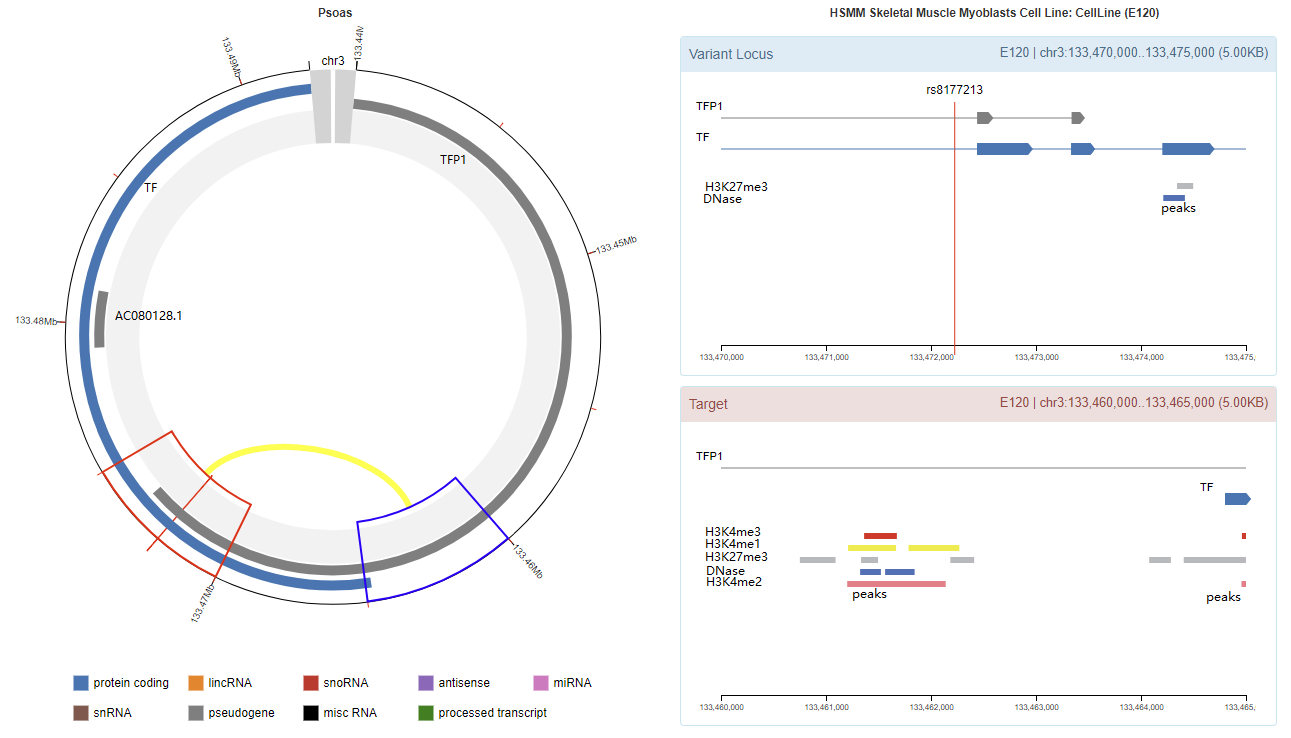
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr3 | 133431016 | 133431089 | E067 | -41138 |
| chr3 | 133436424 | 133436504 | E067 | -35723 |
| chr3 | 133461397 | 133461916 | E067 | -10311 |
| chr3 | 133461945 | 133462055 | E067 | -10172 |
| chr3 | 133464069 | 133464119 | E067 | -8108 |
| chr3 | 133464448 | 133464526 | E067 | -7701 |
| chr3 | 133482923 | 133483028 | E067 | 10696 |
| chr3 | 133483054 | 133483594 | E067 | 10827 |
| chr3 | 133483998 | 133484070 | E067 | 11771 |
| chr3 | 133436424 | 133436504 | E068 | -35723 |
| chr3 | 133464069 | 133464119 | E068 | -8108 |
| chr3 | 133482562 | 133482616 | E068 | 10335 |
| chr3 | 133482923 | 133483028 | E068 | 10696 |
| chr3 | 133483054 | 133483594 | E068 | 10827 |
| chr3 | 133431016 | 133431089 | E069 | -41138 |
| chr3 | 133436424 | 133436504 | E069 | -35723 |
| chr3 | 133461397 | 133461916 | E069 | -10311 |
| chr3 | 133461945 | 133462055 | E069 | -10172 |
| chr3 | 133464069 | 133464119 | E069 | -8108 |
| chr3 | 133473014 | 133473073 | E069 | 787 |
| chr3 | 133473315 | 133473659 | E069 | 1088 |
| chr3 | 133476260 | 133476458 | E069 | 4033 |
| chr3 | 133482562 | 133482616 | E069 | 10335 |
| chr3 | 133482923 | 133483028 | E069 | 10696 |
| chr3 | 133483054 | 133483594 | E069 | 10827 |
| chr3 | 133483998 | 133484070 | E069 | 11771 |
| chr3 | 133484337 | 133484387 | E069 | 12110 |
| chr3 | 133482923 | 133483028 | E070 | 10696 |
| chr3 | 133483054 | 133483594 | E070 | 10827 |
| chr3 | 133431016 | 133431089 | E071 | -41138 |
| chr3 | 133436424 | 133436504 | E071 | -35723 |
| chr3 | 133461397 | 133461916 | E071 | -10311 |
| chr3 | 133461945 | 133462055 | E071 | -10172 |
| chr3 | 133464069 | 133464119 | E071 | -8108 |
| chr3 | 133473014 | 133473073 | E071 | 787 |
| chr3 | 133473315 | 133473659 | E071 | 1088 |
| chr3 | 133482562 | 133482616 | E071 | 10335 |
| chr3 | 133482923 | 133483028 | E071 | 10696 |
| chr3 | 133483054 | 133483594 | E071 | 10827 |
| chr3 | 133483998 | 133484070 | E071 | 11771 |
| chr3 | 133484337 | 133484387 | E071 | 12110 |
| chr3 | 133431016 | 133431089 | E072 | -41138 |
| chr3 | 133461397 | 133461916 | E072 | -10311 |
| chr3 | 133461945 | 133462055 | E072 | -10172 |
| chr3 | 133464069 | 133464119 | E072 | -8108 |
| chr3 | 133464448 | 133464526 | E072 | -7701 |
| chr3 | 133473014 | 133473073 | E072 | 787 |
| chr3 | 133482923 | 133483028 | E072 | 10696 |
| chr3 | 133483054 | 133483594 | E072 | 10827 |
| chr3 | 133483998 | 133484070 | E072 | 11771 |
| chr3 | 133484337 | 133484387 | E072 | 12110 |
| chr3 | 133436424 | 133436504 | E073 | -35723 |
| chr3 | 133461397 | 133461916 | E073 | -10311 |
| chr3 | 133461945 | 133462055 | E073 | -10172 |
| chr3 | 133464448 | 133464526 | E073 | -7701 |
| chr3 | 133482923 | 133483028 | E073 | 10696 |
| chr3 | 133483054 | 133483594 | E073 | 10827 |
| chr3 | 133431016 | 133431089 | E074 | -41138 |
| chr3 | 133436424 | 133436504 | E074 | -35723 |
| chr3 | 133461397 | 133461916 | E074 | -10311 |
| chr3 | 133461945 | 133462055 | E074 | -10172 |
| chr3 | 133464069 | 133464119 | E074 | -8108 |
| chr3 | 133473014 | 133473073 | E074 | 787 |
| chr3 | 133473315 | 133473659 | E074 | 1088 |
| chr3 | 133476260 | 133476458 | E074 | 4033 |
| chr3 | 133482562 | 133482616 | E074 | 10335 |
| chr3 | 133482923 | 133483028 | E074 | 10696 |
| chr3 | 133483054 | 133483594 | E074 | 10827 |
| chr3 | 133483998 | 133484070 | E074 | 11771 |
| chr3 | 133484337 | 133484387 | E074 | 12110 |
| chr3 | 133464448 | 133464526 | E082 | -7701 |
Promoter Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance(-/+:Up/Downstream) |
|---|---|---|---|---|
| chr3 | 133464975 | 133465152 | E067 | -7075 |
| chr3 | 133465195 | 133465439 | E067 | -6788 |
| chr3 | 133465691 | 133465761 | E067 | -6466 |
| chr3 | 133468272 | 133468322 | E067 | -3905 |
| chr3 | 133464975 | 133465152 | E068 | -7075 |
| chr3 | 133465195 | 133465439 | E068 | -6788 |
| chr3 | 133465691 | 133465761 | E068 | -6466 |
| chr3 | 133468272 | 133468322 | E068 | -3905 |
| chr3 | 133464975 | 133465152 | E069 | -7075 |
| chr3 | 133465195 | 133465439 | E069 | -6788 |
| chr3 | 133465691 | 133465761 | E069 | -6466 |
| chr3 | 133468272 | 133468322 | E069 | -3905 |
| chr3 | 133465195 | 133465439 | E070 | -6788 |
| chr3 | 133464975 | 133465152 | E071 | -7075 |
| chr3 | 133465195 | 133465439 | E071 | -6788 |
| chr3 | 133465691 | 133465761 | E071 | -6466 |
| chr3 | 133468272 | 133468322 | E071 | -3905 |
| chr3 | 133464975 | 133465152 | E072 | -7075 |
| chr3 | 133465195 | 133465439 | E072 | -6788 |
| chr3 | 133465691 | 133465761 | E072 | -6466 |
| chr3 | 133468272 | 133468322 | E072 | -3905 |
| chr3 | 133464975 | 133465152 | E073 | -7075 |
| chr3 | 133465195 | 133465439 | E073 | -6788 |
| chr3 | 133465691 | 133465761 | E073 | -6466 |
| chr3 | 133468272 | 133468322 | E073 | -3905 |
| chr3 | 133464975 | 133465152 | E074 | -7075 |
| chr3 | 133465195 | 133465439 | E074 | -6788 |
| chr3 | 133465691 | 133465761 | E074 | -6466 |
| chr3 | 133468272 | 133468322 | E074 | -3905 |
| chr3 | 133464975 | 133465152 | E081 | -7075 |
| chr3 | 133464975 | 133465152 | E082 | -7075 |
| chr3 | 133465195 | 133465439 | E082 | -6788 |