rs9508066
Homo sapiens
G>A
LOC105370135 : Intron Variant
SNV (Single Nucleotide Variation)
A=0314 (9403/29856,GnomAD)
A=0293 (8544/29116,TOPMED)
A=0267 (1336/5008,1000G)
A=0304 (1173/3854,ALSPAC)
A=0289 (1072/3708,TWINSUK)
A=0293 (8544/29116,TOPMED)
A=0267 (1336/5008,1000G)
A=0304 (1173/3854,ALSPAC)
A=0289 (1072/3708,TWINSUK)
chr13:28578368 (GRCh38.p7) (13q12.3)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 13 | NC_000013.11:g.28578368G>A |
| GRCh37.p13 chr 13 | NC_000013.10:g.29152505G>A |
Gene: LOC105370135, uncharacterized LOC105370135(plus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| LOC105370135 transcript | XR_941799.2:n. | N/A | Intron Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | G=0.694 | A=0.306 |
| 1000Genomes | American | Sub | 694 | G=0.850 | A=0.150 |
| 1000Genomes | East Asian | Sub | 1008 | G=0.709 | A=0.291 |
| 1000Genomes | Europe | Sub | 1006 | G=0.716 | A=0.284 |
| 1000Genomes | Global | Study-wide | 5008 | G=0.733 | A=0.267 |
| 1000Genomes | South Asian | Sub | 978 | G=0.750 | A=0.250 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | G=0.696 | A=0.304 |
| The Genome Aggregation Database | African | Sub | 8676 | G=0.683 | A=0.317 |
| The Genome Aggregation Database | American | Sub | 838 | G=0.820 | A=0.180 |
| The Genome Aggregation Database | East Asian | Sub | 1610 | G=0.753 | A=0.247 |
| The Genome Aggregation Database | Europe | Sub | 18430 | G=0.674 | A=0.325 |
| The Genome Aggregation Database | Global | Study-wide | 29856 | G=0.685 | A=0.314 |
| The Genome Aggregation Database | Other | Sub | 302 | G=0.650 | A=0.350 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29116 | G=0.706 | A=0.293 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | G=0.711 | A=0.289 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 21314694 | Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. | Kendler KS | Alcohol Clin Exp Res |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs9508066 | 0.000643 | alcohol dependence | 21314694 |
eQTL of rs9508066 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs9508066 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
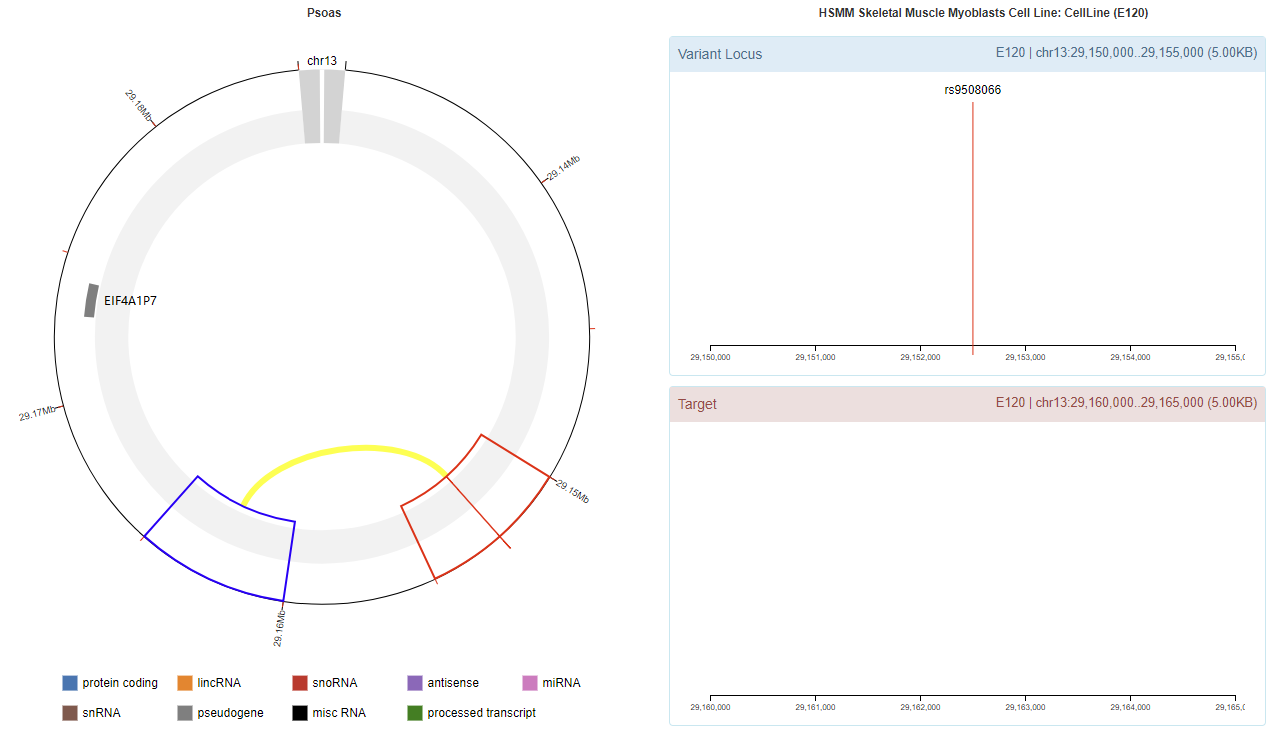
Genomic View
Chromatin Interaction
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr13 | 29142024 | 29142159 | E067 | -10346 |
| chr13 | 29142244 | 29142924 | E067 | -9581 |
| chr13 | 29174776 | 29174826 | E067 | 22271 |
| chr13 | 29175150 | 29175200 | E067 | 22645 |
| chr13 | 29175468 | 29175518 | E067 | 22963 |
| chr13 | 29175821 | 29176147 | E067 | 23316 |
| chr13 | 29176591 | 29176641 | E067 | 24086 |
| chr13 | 29177086 | 29177186 | E067 | 24581 |
| chr13 | 29142244 | 29142924 | E068 | -9581 |
| chr13 | 29175150 | 29175200 | E068 | 22645 |
| chr13 | 29175468 | 29175518 | E068 | 22963 |
| chr13 | 29175821 | 29176147 | E068 | 23316 |
| chr13 | 29142024 | 29142159 | E069 | -10346 |
| chr13 | 29142244 | 29142924 | E069 | -9581 |
| chr13 | 29175821 | 29176147 | E069 | 23316 |
| chr13 | 29176591 | 29176641 | E069 | 24086 |
| chr13 | 29104665 | 29104756 | E070 | -47749 |
| chr13 | 29104665 | 29104756 | E071 | -47749 |
| chr13 | 29142024 | 29142159 | E071 | -10346 |
| chr13 | 29142244 | 29142924 | E071 | -9581 |
| chr13 | 29175468 | 29175518 | E071 | 22963 |
| chr13 | 29175821 | 29176147 | E071 | 23316 |
| chr13 | 29177086 | 29177186 | E071 | 24581 |
| chr13 | 29177310 | 29177435 | E071 | 24805 |
| chr13 | 29177473 | 29177644 | E071 | 24968 |
| chr13 | 29142024 | 29142159 | E072 | -10346 |
| chr13 | 29142244 | 29142924 | E072 | -9581 |
| chr13 | 29174776 | 29174826 | E072 | 22271 |
| chr13 | 29175150 | 29175200 | E072 | 22645 |
| chr13 | 29175468 | 29175518 | E072 | 22963 |
| chr13 | 29175821 | 29176147 | E072 | 23316 |
| chr13 | 29176591 | 29176641 | E072 | 24086 |
| chr13 | 29142244 | 29142924 | E073 | -9581 |
| chr13 | 29104665 | 29104756 | E074 | -47749 |
| chr13 | 29131964 | 29132615 | E074 | -19890 |
| chr13 | 29132733 | 29133095 | E074 | -19410 |
| chr13 | 29142024 | 29142159 | E074 | -10346 |
| chr13 | 29142244 | 29142924 | E074 | -9581 |
| chr13 | 29175150 | 29175200 | E074 | 22645 |
| chr13 | 29175468 | 29175518 | E074 | 22963 |
| chr13 | 29175821 | 29176147 | E074 | 23316 |
| chr13 | 29176591 | 29176641 | E074 | 24086 |
| chr13 | 29177086 | 29177186 | E074 | 24581 |
| chr13 | 29104665 | 29104756 | E081 | -47749 |
Promoter Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance(-/+:Up/Downstream) |
|---|---|---|---|---|
| chr13 | 29104909 | 29107742 | E067 | -44763 |
| chr13 | 29104909 | 29107742 | E068 | -44763 |
| chr13 | 29104909 | 29107742 | E071 | -44763 |
| chr13 | 29104909 | 29107742 | E072 | -44763 |
| chr13 | 29104909 | 29107742 | E073 | -44763 |
| chr13 | 29104909 | 29107742 | E074 | -44763 |
| chr13 | 29104909 | 29107742 | E082 | -44763 |