rs964170
Homo sapiens
G>T
ARHGAP10 : Intron Variant
SNV (Single Nucleotide Variation)
G==0246 (7379/29918,GnomAD)
G==0363 (10570/29118,TOPMED)
G==0320 (1601/5008,1000G)
G==0058 (225/3854,ALSPAC)
G==0055 (204/3708,TWINSUK)
G==0363 (10570/29118,TOPMED)
G==0320 (1601/5008,1000G)
G==0058 (225/3854,ALSPAC)
G==0055 (204/3708,TWINSUK)
chr4:147774805 (GRCh38.p7) (4q31.23)
ND
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 4 | NC_000004.12:g.147774805G>T |
| GRCh37.p13 chr 4 | NC_000004.11:g.148695956G>T |
Gene: ARHGAP10, Rho GTPase activating protein 10(plus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| ARHGAP10 transcript | NM_024605.3:c. | N/A | Intron Variant |
| ARHGAP10 transcript variant X1 | XM_005263215.3:c. | N/A | Intron Variant |
| ARHGAP10 transcript variant X2 | XM_017008602.1:c. | N/A | Intron Variant |
| ARHGAP10 transcript variant X3 | XR_001741324.1:n. | N/A | Intron Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | G=0.821 | T=0.179 |
| 1000Genomes | American | Sub | 694 | G=0.140 | T=0.860 |
| 1000Genomes | East Asian | Sub | 1008 | G=0.147 | T=0.853 |
| 1000Genomes | Europe | Sub | 1006 | G=0.063 | T=0.937 |
| 1000Genomes | Global | Study-wide | 5008 | G=0.320 | T=0.680 |
| 1000Genomes | South Asian | Sub | 978 | G=0.210 | T=0.790 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | G=0.058 | T=0.942 |
| The Genome Aggregation Database | African | Sub | 8694 | G=0.694 | T=0.306 |
| The Genome Aggregation Database | American | Sub | 836 | G=0.140 | T=0.860 |
| The Genome Aggregation Database | East Asian | Sub | 1614 | G=0.136 | T=0.864 |
| The Genome Aggregation Database | Europe | Sub | 18472 | G=0.053 | T=0.946 |
| The Genome Aggregation Database | Global | Study-wide | 29918 | G=0.246 | T=0.753 |
| The Genome Aggregation Database | Other | Sub | 302 | G=0.090 | T=0.910 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | G=0.363 | T=0.637 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | G=0.055 | T=0.945 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 20158304 | A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. | Lind PA | Twin Res Hum Genet |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs964170 | 4.43E-08 | Nicotine dependence (smoking) | 20158304 |
| rs964170 | 0.00000149 | Comorbid alcohol and nicotine dependence | 20158304 |
eQTL of rs964170 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs964170 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
Genomic View
Chromatin Interaction
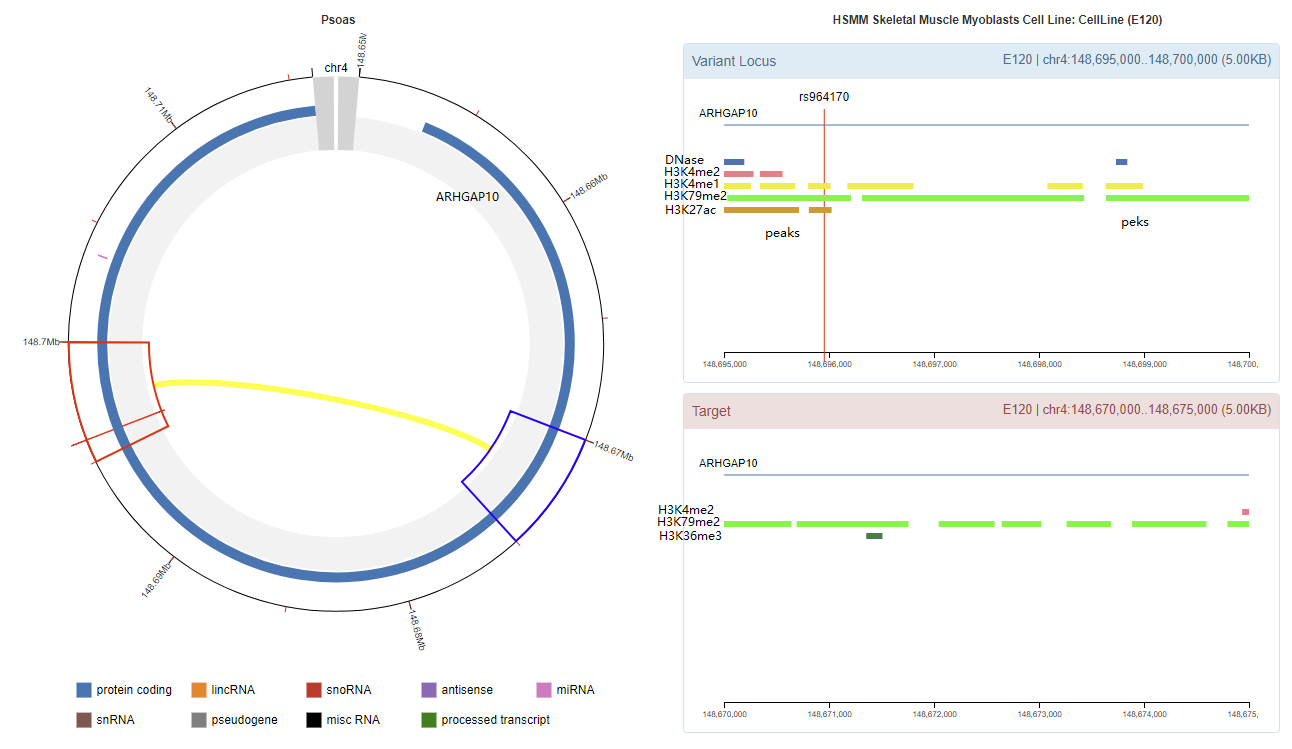
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr4 | 148676713 | 148677287 | E067 | -18669 |
| chr4 | 148706298 | 148706613 | E068 | 10342 |
| chr4 | 148706684 | 148706904 | E068 | 10728 |
| chr4 | 148729292 | 148729365 | E068 | 33336 |
| chr4 | 148729728 | 148729817 | E068 | 33772 |
| chr4 | 148737655 | 148737815 | E068 | 41699 |
| chr4 | 148737876 | 148737949 | E068 | 41920 |
| chr4 | 148737953 | 148738016 | E068 | 41997 |
| chr4 | 148738054 | 148738175 | E068 | 42098 |
| chr4 | 148742192 | 148742577 | E068 | 46236 |
| chr4 | 148742781 | 148743119 | E068 | 46825 |
| chr4 | 148743153 | 148743973 | E068 | 47197 |
| chr4 | 148671037 | 148671124 | E069 | -24832 |
| chr4 | 148671037 | 148671124 | E070 | -24832 |
| chr4 | 148671290 | 148671358 | E070 | -24598 |
| chr4 | 148671398 | 148671460 | E070 | -24496 |
| chr4 | 148723052 | 148723261 | E070 | 27096 |
| chr4 | 148723387 | 148723442 | E070 | 27431 |
| chr4 | 148677893 | 148678008 | E071 | -17948 |
| chr4 | 148706298 | 148706613 | E071 | 10342 |
| chr4 | 148706684 | 148706904 | E071 | 10728 |
| chr4 | 148734663 | 148734876 | E071 | 38707 |
| chr4 | 148737080 | 148737197 | E071 | 41124 |
| chr4 | 148675140 | 148675617 | E072 | -20339 |
| chr4 | 148706298 | 148706613 | E072 | 10342 |
| chr4 | 148706684 | 148706904 | E072 | 10728 |
| chr4 | 148675140 | 148675617 | E073 | -20339 |
| chr4 | 148676713 | 148677287 | E073 | -18669 |
| chr4 | 148701591 | 148701723 | E073 | 5635 |
| chr4 | 148701746 | 148701933 | E073 | 5790 |
| chr4 | 148671037 | 148671124 | E081 | -24832 |
| chr4 | 148671290 | 148671358 | E081 | -24598 |
| chr4 | 148671398 | 148671460 | E081 | -24496 |
Promoter Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance(-/+:Up/Downstream) |
|---|---|---|---|---|
| chr4 | 148652166 | 148654713 | E067 | -41243 |
| chr4 | 148652166 | 148654713 | E068 | -41243 |
| chr4 | 148652166 | 148654713 | E069 | -41243 |
| chr4 | 148652166 | 148654713 | E071 | -41243 |
| chr4 | 148652166 | 148654713 | E072 | -41243 |
| chr4 | 148652166 | 148654713 | E073 | -41243 |
| chr4 | 148652166 | 148654713 | E074 | -41243 |
| chr4 | 148652166 | 148654713 | E082 | -41243 |