PAN3
Entrez ID: 255967
Full name: poly(A) specific ribonuclease subunit PAN3
External links: HGNC UniprotKB Ensembl RefSeq COSMIC
Family: Other: PAN3
Chromosomal location: 13q12.2
Substructure location: Gatekeeper: A-loop: G-loop: αC-helix:
Please click HERE
for better display.
PAN3
Regulatory subunit of the poly(A)-nuclease (PAN) deadenylation complex, one of two cytoplasmic mRNA deadenylases involved in general and miRNA-mediated mRNA turnover. PAN specifically shortens poly(A) tails of RNA and the activity is stimulated by poly(A)-binding protein (PABP). PAN deadenylation is...
Regulatory subunit of the poly(A)-nuclease (PAN) deadenylation complex, one of two cytoplasmic mRNA deadenylases involved in general and miRNA-mediated mRNA turnover. PAN specifically shortens poly(A) tails of RNA and the activity is stimulated by poly(A)-binding protein (PABP). PAN deadenylation is followed by rapid degradation of the shortened mRNA tails by the CCR4-NOT complex. Deadenylated mRNAs are then degraded by two alternative mechanisms, namely exosome-mediated 3'-5' exonucleolytic degradation, or deadenlyation-dependent mRNA decaping and subsequent 5'-3' exonucleolytic degradation by XRN1. PAN3 acts as a positive regulator for PAN activity, recruiting the catalytic subunit PAN2 to mRNA via its interaction with RNA and PABP, and to miRNA targets via its interaction with GW182 family proteins.
View more >>
GO - Biological processes (BP):
deadenylation-dependent decapping of nuclear-transcribed mRNA, mRNA processing, nuclear-transcribed mRNA poly(A) tail shortening, positive regulation of cytoplasmic mRNA processing body assembly, protein targeting, RNA phosphodiester bond hydrolysis, exonucleolytic...
deadenylation-dependent decapping of nuclear-transcribed mRNA, mRNA processing, nuclear-transcribed mRNA poly(A) tail shortening, positive regulation of cytoplasmic mRNA processing body assembly, protein targeting, RNA phosphodiester bond hydrolysis, exonucleolytic
View more >>
GO - Molecular function (MF):
ATP binding, metal ion binding, protein kinase activity, RNA binding...
ATP binding, metal ion binding, protein kinase activity, RNA binding
View more >>
GO - Cellular component (CC):
cytosol, P-body, PAN complex...
cytosol, P-body, PAN complex
View more >>
poly(A) specific ribonuclease subunit PAN3
Control panel
Show dataset:
Show substructure:
All sites
Gatekeeper
A-loop
G-loop
αC-helix
Gatekeeper
A-loop
G-loop
αC-helix
Filter mutation sites
All sites
Minimum 5 samples
Minimum 10 samples
Minimum 5 samples
Minimum 10 samples
Data table is loading....
Kaplan plot for PAN3 in
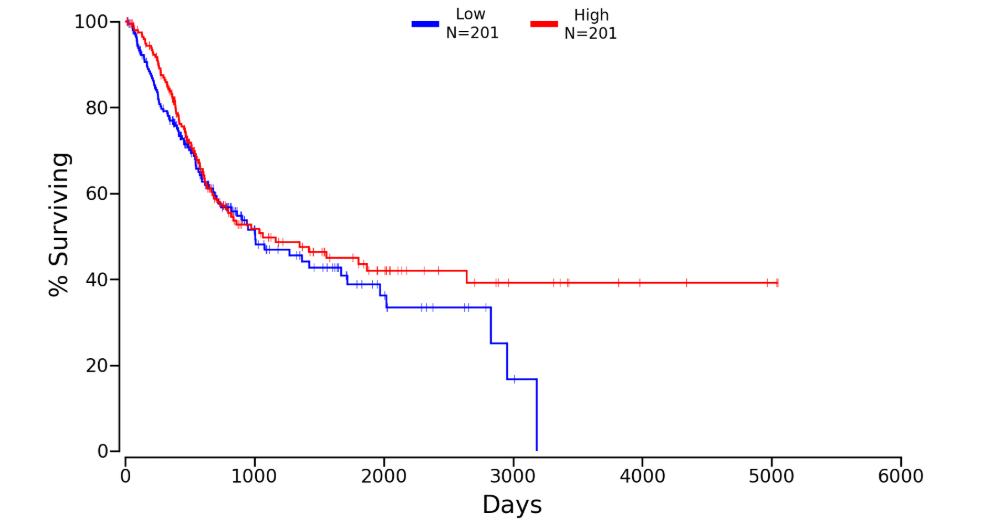
Download patient group data (Lower:Upper = 50%:50%)
*NOTE: the Kaplan plots were collected from OncoLnc