SRPK1
Entrez ID: 6732
Full name: SRSF protein kinase 1
External links: HGNC UniprotKB Ensembl RefSeq COSMIC
Family: CMGC: SRPK
Chromosomal location: 6p21.31
Substructure location: Gatekeeper: 165 A-loop: 496...516 G-loop: [N/A] αC-helix: 115...133
Please click HERE
for better display.
SRPK1
Serine/arginine-rich protein-specific kinase which specifically phosphorylates its substrates at serine residues located in regions rich in arginine/serine dipeptides, known as RS domains and is involved in the phosphorylation of SR splicing factors and the regulation of splicing. Plays a central ro...
Serine/arginine-rich protein-specific kinase which specifically phosphorylates its substrates at serine residues located in regions rich in arginine/serine dipeptides, known as RS domains and is involved in the phosphorylation of SR splicing factors and the regulation of splicing. Plays a central role in the regulatory network for splicing, controlling the intranuclear distribution of splicing factors in interphase cells and the reorganization of nuclear speckles during mitosis. Can influence additional steps of mRNA maturation, as well as other cellular activities, such as chromatin reorganization in somatic and sperm cells and cell cycle progression. Isoform 2 phosphorylates SFRS2, ZRSR2, LBR and PRM1. Isoform 2 phosphorylates SRSF1 using a directional (C-terminal to N-terminal) and a dual-track mechanism incorporating both processive phosphorylation (in which the kinase stays attached to the substrate after each round of phosphorylation) and distributive phosphorylation steps (in which the kinase and substrate dissociate after each phosphorylation event). The RS domain of SRSF1 binds first to a docking groove in the large lobe of the kinase domain of SRPK1. This induces certain structural changes in SRPK1 and/or RRM2 domain of SRSF1, allowing RRM2 to bind the kinase and initiate phosphorylation. The cycles continue for several phosphorylation steps in a processive manner (steps 1-8) until the last few phosphorylation steps (approximately steps 9-12). During that time, a mechanical stress induces the unfolding of the beta-4 motif in RRM2, which then docks at the docking groove of SRPK1. This also signals RRM2 to begin to dissociate, which facilitates SRSF1 dissociation after phosphorylation is completed. Isoform 2 can mediate hepatitis B virus (HBV) core protein phosphorylation. It plays a negative role in the regulation of HBV replication through a mechanism not involving the phosphorylation of the core protein but by reducing the packaging efficiency of the pregenomic RNA (pgRNA) without affecting the formation of the viral core particles. Isoform 1 and isoform 2 can induce splicing of exon 10 in MAPT/TAU. The ratio of isoform 1/isoform 2 plays a decisive role in determining cell fate in K-562 leukaemic cell line: isoform 2 favors proliferation where as isoform 1 favors differentiation.
View more >>
GO - Biological processes (BP):
chromosome segregation, innate immune response, intracellular signal transduction, negative regulation of viral genome replication, positive regulation of viral genome replication, protein phosphorylation, regulation of gene expression, regulation of mRNA processing, regulation of mRNA splicing, via...
chromosome segregation, innate immune response, intracellular signal transduction, negative regulation of viral genome replication, positive regulation of viral genome replication, protein phosphorylation, regulation of gene expression, regulation of mRNA processing, regulation of mRNA splicing, via spliceosome, RNA splicing, sperm chromatin condensation, spliceosomal complex assembly, viral process
View more >>
GO - Molecular function (MF):
ATP binding, magnesium ion binding, protein kinase activity, protein serine/threonine kinase activity, RNA binding...
ATP binding, magnesium ion binding, protein kinase activity, protein serine/threonine kinase activity, RNA binding
View more >>
GO - Cellular component (CC):
cytosol, endoplasmic reticulum, nuclear matrix, nucleoplasm, nucleus, plasma membrane, cytoplasm...
cytosol, endoplasmic reticulum, nuclear matrix, nucleoplasm, nucleus, plasma membrane, cytoplasm
View more >>
SRSF protein kinase 1
Control panel
Show dataset:
Show substructure:
All sites
Gatekeeper
A-loop
G-loop
αC-helix
Gatekeeper
A-loop
G-loop
αC-helix
Filter mutation sites
All sites
Minimum 5 samples
Minimum 10 samples
Minimum 5 samples
Minimum 10 samples
Data table is loading....
Kaplan plot for SRPK1 in
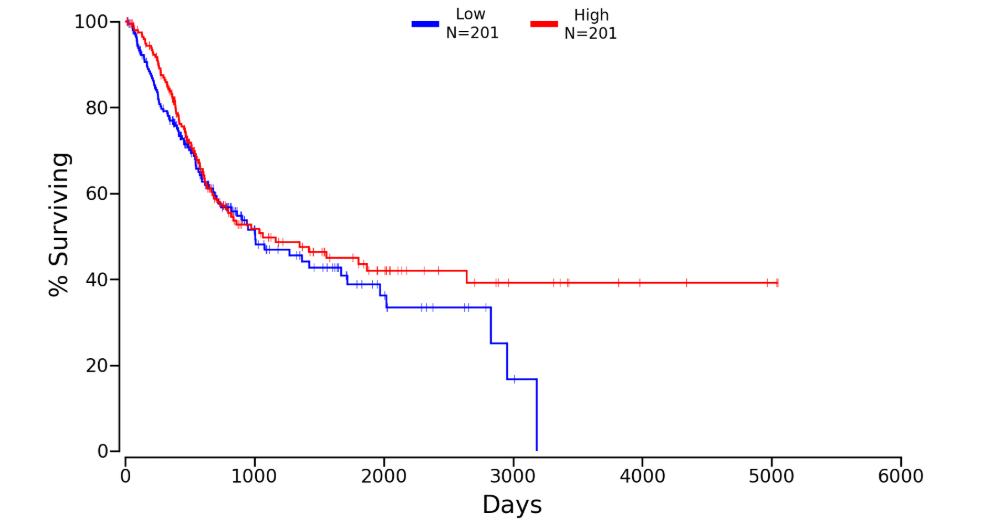
Download patient group data (Lower:Upper = 50%:50%)
*NOTE: the Kaplan plots were collected from OncoLnc