Gene Page: TXNRD2
Summary ?
| GeneID | 10587 |
| Symbol | TXNRD2 |
| Synonyms | SELZ|TR|TR-BETA|TR3|TRXR2 |
| Description | thioredoxin reductase 2 |
| Reference | MIM:606448|HGNC:HGNC:18155|Ensembl:ENSG00000184470|HPRD:05921|HPRD:18519|Vega:OTTHUMG00000149975 |
| Gene type | protein-coding |
| Map location | 22q11.21 |
| Pascal p-value | 0.416 |
| Sherlock p-value | 0.032 |
| Fetal beta | -0.893 |
| eGene | Cerebellum Cortex Frontal Cortex BA9 Hippocampus Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs35374520 | 22 | 19851149 | TXNRD2 | ENSG00000184470.15 | 1.453E-6 | 0.01 | 69900 | gtex_brain_putamen_basal |
| rs13053818 | 22 | 19859652 | TXNRD2 | ENSG00000184470.15 | 2.992E-7 | 0.01 | 61397 | gtex_brain_putamen_basal |
| rs13056219 | 22 | 19860846 | TXNRD2 | ENSG00000184470.15 | 3.012E-7 | 0.01 | 60203 | gtex_brain_putamen_basal |
| rs7290770 | 22 | 19861161 | TXNRD2 | ENSG00000184470.15 | 3.294E-7 | 0.01 | 59888 | gtex_brain_putamen_basal |
| rs1044732 | 22 | 19863142 | TXNRD2 | ENSG00000184470.15 | 3.012E-7 | 0.01 | 57907 | gtex_brain_putamen_basal |
| rs16984299 | 22 | 19863593 | TXNRD2 | ENSG00000184470.15 | 1.708E-6 | 0.01 | 57456 | gtex_brain_putamen_basal |
| rs7288061 | 22 | 19864239 | TXNRD2 | ENSG00000184470.15 | 3.012E-7 | 0.01 | 56810 | gtex_brain_putamen_basal |
| rs7288170 | 22 | 19864252 | TXNRD2 | ENSG00000184470.15 | 2.298E-6 | 0.01 | 56797 | gtex_brain_putamen_basal |
| rs7287073 | 22 | 19864335 | TXNRD2 | ENSG00000184470.15 | 3.012E-7 | 0.01 | 56714 | gtex_brain_putamen_basal |
| rs5993849 | 22 | 19866194 | TXNRD2 | ENSG00000184470.15 | 1.379E-6 | 0.01 | 54855 | gtex_brain_putamen_basal |
| rs34175429 | 22 | 19867276 | TXNRD2 | ENSG00000184470.15 | 2.377E-6 | 0.01 | 53773 | gtex_brain_putamen_basal |
| rs1139795 | 22 | 19867771 | TXNRD2 | ENSG00000184470.15 | 9.454E-7 | 0.01 | 53278 | gtex_brain_putamen_basal |
| rs13054713 | 22 | 19867914 | TXNRD2 | ENSG00000184470.15 | 9.121E-8 | 0.01 | 53135 | gtex_brain_putamen_basal |
| rs397897683 | 22 | 19868255 | TXNRD2 | ENSG00000184470.15 | 6.193E-7 | 0.01 | 52794 | gtex_brain_putamen_basal |
| rs45465601 | 22 | 19868678 | TXNRD2 | ENSG00000184470.15 | 9.454E-7 | 0.01 | 52371 | gtex_brain_putamen_basal |
| rs34318203 | 22 | 19868925 | TXNRD2 | ENSG00000184470.15 | 1.157E-6 | 0.01 | 52124 | gtex_brain_putamen_basal |
| rs33997573 | 22 | 19868951 | TXNRD2 | ENSG00000184470.15 | 9.378E-8 | 0.01 | 52098 | gtex_brain_putamen_basal |
| rs34820460 | 22 | 19869210 | TXNRD2 | ENSG00000184470.15 | 6.716E-7 | 0.01 | 51839 | gtex_brain_putamen_basal |
| rs5993850 | 22 | 19869703 | TXNRD2 | ENSG00000184470.15 | 9.946E-7 | 0.01 | 51346 | gtex_brain_putamen_basal |
| rs8141451 | 22 | 19870036 | TXNRD2 | ENSG00000184470.15 | 9.983E-7 | 0.01 | 51013 | gtex_brain_putamen_basal |
| rs8141610 | 22 | 19870147 | TXNRD2 | ENSG00000184470.15 | 1.696E-7 | 0.01 | 50902 | gtex_brain_putamen_basal |
| rs5993851 | 22 | 19871553 | TXNRD2 | ENSG00000184470.15 | 4.563E-7 | 0.01 | 49496 | gtex_brain_putamen_basal |
| rs12158214 | 22 | 19871691 | TXNRD2 | ENSG00000184470.15 | 2.889E-7 | 0.01 | 49358 | gtex_brain_putamen_basal |
| rs34249993 | 22 | 19872170 | TXNRD2 | ENSG00000184470.15 | 5.996E-7 | 0.01 | 48879 | gtex_brain_putamen_basal |
| rs73148965 | 22 | 19872935 | TXNRD2 | ENSG00000184470.15 | 4.45E-7 | 0.01 | 48114 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
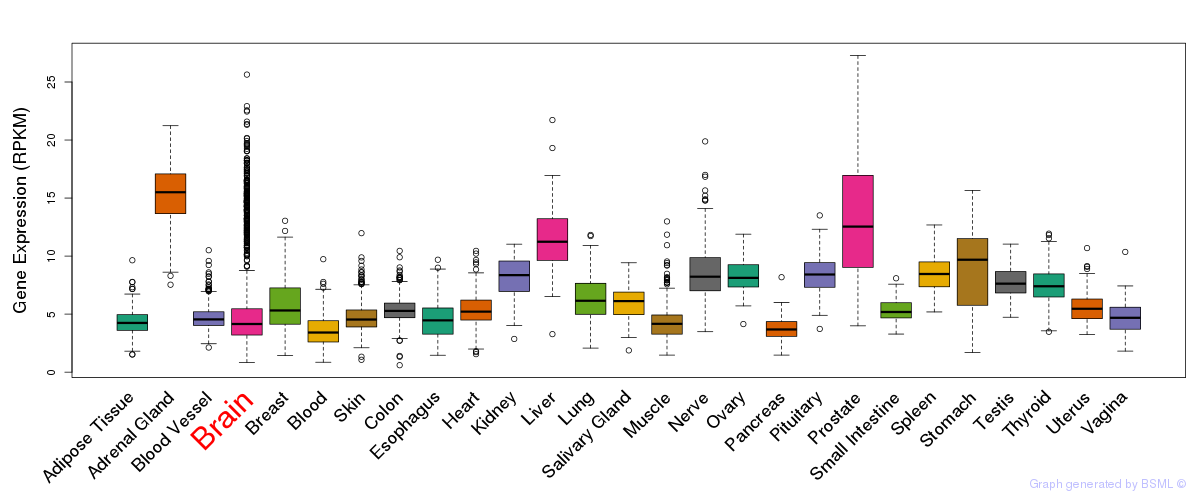
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CAPRIN2 | 0.82 | 0.78 |
| AC008443.1 | 0.78 | 0.74 |
| UGCGL2 | 0.77 | 0.64 |
| CCDC66 | 0.76 | 0.63 |
| TTC14 | 0.76 | 0.65 |
| PPWD1 | 0.75 | 0.71 |
| WSB1 | 0.75 | 0.64 |
| AC008806.1 | 0.74 | 0.81 |
| ZNF382 | 0.74 | 0.74 |
| SFRS5 | 0.74 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.52 | -0.56 |
| MT-CO2 | -0.51 | -0.54 |
| AF347015.21 | -0.50 | -0.55 |
| HIGD1B | -0.50 | -0.56 |
| AF347015.2 | -0.49 | -0.51 |
| AF347015.8 | -0.49 | -0.52 |
| MT-CYB | -0.49 | -0.51 |
| AF347015.33 | -0.48 | -0.49 |
| IFI27 | -0.48 | -0.54 |
| AF347015.27 | -0.48 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004791 | thioredoxin-disulfide reductase activity | ISS | - | |
| GO:0008430 | selenium binding | IEA | - | |
| GO:0016654 | oxidoreductase activity, acting on NADH or NADPH, disulfide as acceptor | IEA | - | |
| GO:0009055 | electron carrier activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0050660 | FAD binding | IEA | - | |
| GO:0050661 | NADP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000305 | response to oxygen radical | TAS | 10455115 | |
| GO:0045454 | cell redox homeostasis | IEA | - | |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | IDA | 10215850 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PYRIMIDINE METABOLISM | 98 | 53 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS | 114 | 71 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| STARK HYPPOCAMPUS 22Q11 DELETION DN | 20 | 19 | All SZGR 2.0 genes in this pathway |
| HOUSTIS ROS | 36 | 29 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD DN | 162 | 102 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE NOT BY 4NQO IN WS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE UP | 56 | 41 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |