Gene Page: CHRNA7
Summary ?
| GeneID | 1139 |
| Symbol | CHRNA7 |
| Synonyms | CHRNA7-2|NACHRA7 |
| Description | cholinergic receptor nicotinic alpha 7 subunit |
| Reference | MIM:118511|HGNC:HGNC:1960|Ensembl:ENSG00000175344|HPRD:00339|Vega:OTTHUMG00000129285 |
| Gene type | protein-coding |
| Map location | 15q14 |
| Pascal p-value | 0.133 |
| Fetal beta | 0.528 |
| Support | LIGAND GATED ION SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 6 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 31 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
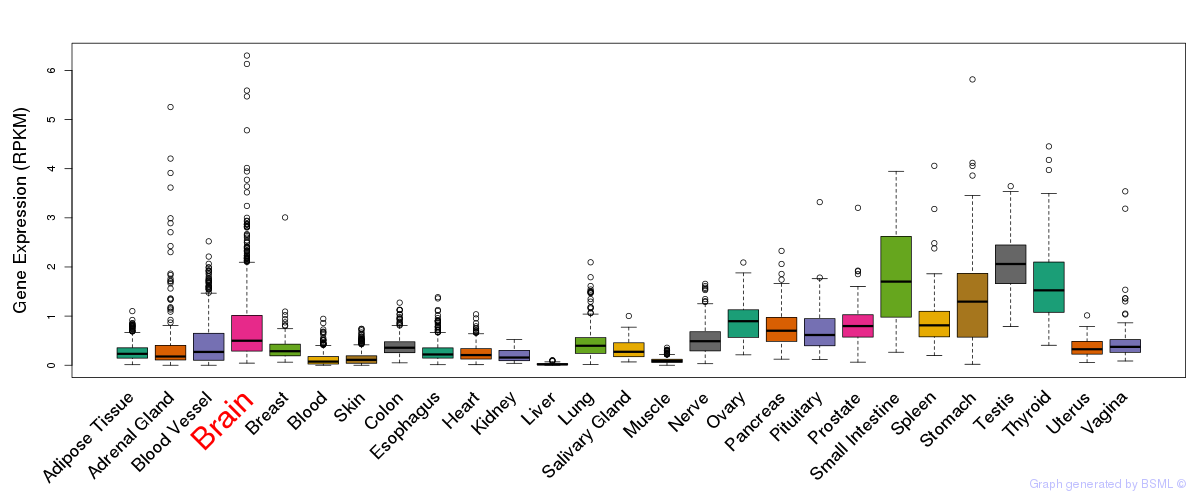
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA0196 | 0.87 | 0.85 |
| TRIP12 | 0.87 | 0.87 |
| MTMR15 | 0.86 | 0.86 |
| IPO8 | 0.86 | 0.85 |
| KIAA1012 | 0.85 | 0.86 |
| FAM178A | 0.85 | 0.86 |
| NBR1 | 0.85 | 0.84 |
| PTK2 | 0.85 | 0.83 |
| UTP14C | 0.84 | 0.86 |
| RBBP5 | 0.84 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.67 | -0.64 |
| IFI27 | -0.64 | -0.67 |
| IL32 | -0.62 | -0.61 |
| MT-CO2 | -0.62 | -0.62 |
| FXYD1 | -0.61 | -0.63 |
| HIGD1B | -0.60 | -0.63 |
| VAMP5 | -0.60 | -0.60 |
| AF347015.31 | -0.59 | -0.62 |
| CXCL14 | -0.59 | -0.63 |
| CSAG1 | -0.59 | -0.59 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0030594 | neurotransmitter receptor activity | IEA | Neurotransmitter (GO term level: 5) | - |
| GO:0015464 | acetylcholine receptor activity | IDA | Neurotransmitter (GO term level: 6) | 8906617 |
| GO:0001540 | beta-amyloid binding | IPI | 10681545 | |
| GO:0004889 | nicotinic acetylcholine-activated cation-selective channel activity | IDA | 8145738 |8906617 | |
| GO:0004889 | nicotinic acetylcholine-activated cation-selective channel activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0005230 | extracellular ligand-gated ion channel activity | IEA | - | |
| GO:0015643 | toxin binding | IDA | 12508119 | |
| GO:0017081 | chloride channel regulator activity | IDA | 8145738 | |
| GO:0042803 | protein homodimerization activity | IDA | 8145738 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007271 | synaptic transmission, cholinergic | IEA | neuron, Cholinergic, Synap, Neurotransmitter (GO term level: 7) | - |
| GO:0032225 | regulation of synaptic transmission, dopaminergic | IEA | neuron, Synap, Neurotransmitter, dopamine (GO term level: 9) | - |
| GO:0000187 | activation of MAPK activity | IDA | 10771023 | |
| GO:0001666 | response to hypoxia | IDA | 12189247 | |
| GO:0001988 | positive regulation of heart rate in baroreceptor response to decreased systemic arterial blood pressure | IEA | - | |
| GO:0007165 | signal transduction | IDA | 8906617 | |
| GO:0008284 | positive regulation of cell proliferation | IMP | 16280133 |17498763 | |
| GO:0008306 | associative learning | IEA | - | |
| GO:0007613 | memory | IEA | - | |
| GO:0007613 | memory | NAS | 10681545 | |
| GO:0006816 | calcium ion transport | IDA | 8145738 | |
| GO:0006816 | calcium ion transport | IMP | 10771023 | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006811 | ion transport | NAS | 8906617 | |
| GO:0006897 | endocytosis | IEA | - | |
| GO:0006874 | cellular calcium ion homeostasis | IMP | 16280133 | |
| GO:0014061 | regulation of norepinephrine secretion | IEA | - | |
| GO:0042391 | regulation of membrane potential | IEA | - | |
| GO:0032094 | response to food | IEA | - | |
| GO:0042698 | ovulation cycle | IEA | - | |
| GO:0042113 | B cell activation | IEA | - | |
| GO:0042110 | T cell activation | IEA | - | |
| GO:0032720 | negative regulation of tumor necrosis factor production | IEA | - | |
| GO:0032720 | negative regulation of tumor necrosis factor production | IMP | 12508119 | |
| GO:0032715 | negative regulation of interleukin-6 production | IEA | - | |
| GO:0032691 | negative regulation of interleukin-1 beta production | IEA | - | |
| GO:0030317 | sperm motility | IEA | - | |
| GO:0035095 | behavioral response to nicotine | IEA | - | |
| GO:0045766 | positive regulation of angiogenesis | IMP | 12189247 |16280133 | |
| GO:0048149 | behavioral response to ethanol | IEA | - | |
| GO:0050728 | negative regulation of inflammatory response | IDA | 12508119 | |
| GO:0050728 | negative regulation of inflammatory response | IEA | - | |
| GO:0060112 | generation of ovulation cycle rhythm | IEA | - | |
| GO:0050890 | cognition | IMP | 16754836 | |
| GO:0050890 | cognition | NAS | 10681545 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005892 | nicotinic acetylcholine-gated receptor-channel complex | IDA | Neurotransmitter (GO term level: 9) | 8906617 |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005624 | membrane fraction | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0016021 | integral to membrane | NAS | 8145738 |8906617 | |
| GO:0009897 | external side of plasma membrane | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 16280133 | |
| GO:0016324 | apical plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | 16 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | 13 | 12 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION | 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS DN | 97 | 53 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL DN | 214 | 124 | All SZGR 2.0 genes in this pathway |