Gene Page: ADD1
Summary ?
| GeneID | 118 |
| Symbol | ADD1 |
| Synonyms | ADDA |
| Description | adducin 1 |
| Reference | MIM:102680|HGNC:HGNC:243|Ensembl:ENSG00000087274|HPRD:00036|Vega:OTTHUMG00000122080 |
| Gene type | protein-coding |
| Map location | 4p16.3 |
| Pascal p-value | 0.116 |
| Sherlock p-value | 0.048 |
| Fetal beta | 0.432 |
| eGene | Myers' cis & trans |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_mGluR5 CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0024 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1578978 | chr8 | 118196802 | ADD1 | 118 | 0.19 | trans |
Section II. Transcriptome annotation
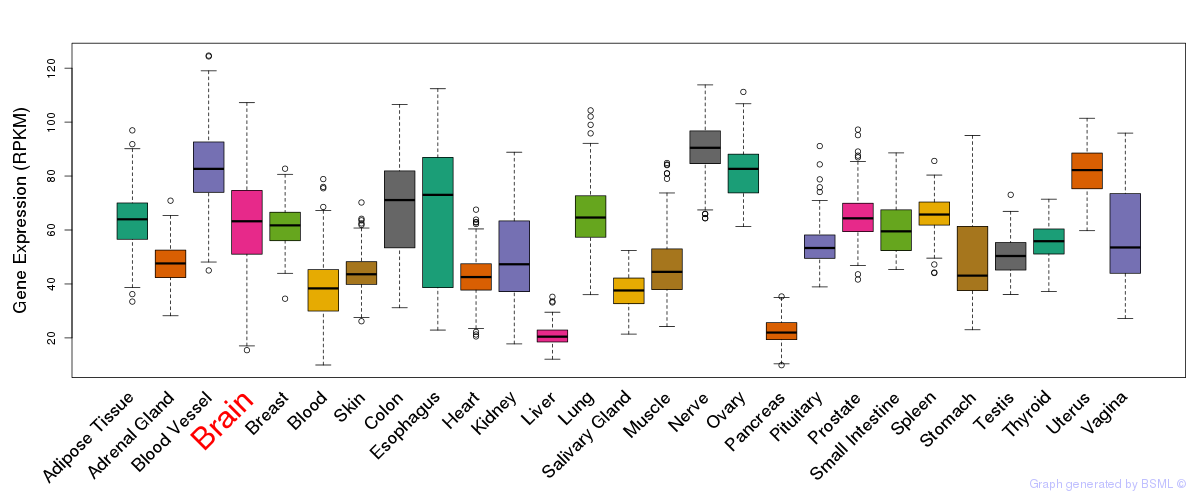
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003779 | actin binding | TAS | 1840603 | |
| GO:0005516 | calmodulin binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10823823 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | 46 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME UNFOLDED PROTEIN RESPONSE | 80 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| WIKMAN ASBESTOS LUNG CANCER UP | 17 | 11 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION UP | 119 | 66 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD UP | 64 | 39 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO BEXAROTENE DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP | 90 | 62 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH DN | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| DAVIES MULTIPLE MYELOMA VS MGUS DN | 28 | 18 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| SYED ESTRADIOL RESPONSE | 19 | 15 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING UP | 93 | 62 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 UP | 115 | 73 | All SZGR 2.0 genes in this pathway |
| STEINER ERYTHROCYTE MEMBRANE GENES | 15 | 10 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-153 | 1384 | 1390 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-299-3p | 1378 | 1385 | 1A,m8 | hsa-miR-299-3p | UAUGUGGGAUGGUAAACCGCUU |
| miR-33 | 90 | 96 | 1A | hsa-miR-33 | GUGCAUUGUAGUUGCAUUG |
| hsa-miR-33b | GUGCAUUGCUGUUGCAUUGCA | ||||
| miR-448 | 1383 | 1390 | 1A,m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.