Gene Page: DLG4
Summary ?
| GeneID | 1742 |
| Symbol | DLG4 |
| Synonyms | PSD95|SAP-90|SAP90 |
| Description | discs large homolog 4 |
| Reference | MIM:602887|HGNC:HGNC:2903|Ensembl:ENSG00000132535|HPRD:04199|Vega:OTTHUMG00000134327 |
| Gene type | protein-coding |
| Map location | 17p13.1 |
| Pascal p-value | 1.301E-4 |
| Fetal beta | -0.465 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | PROTEIN CLUSTERING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_PocklingtonH1 G2Cdb.human_Synaptosome G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 7 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 7.5711 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04106389 | 17 | 7117241 | DLG4 | 2.334E-4 | 0.308 | 0.037 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
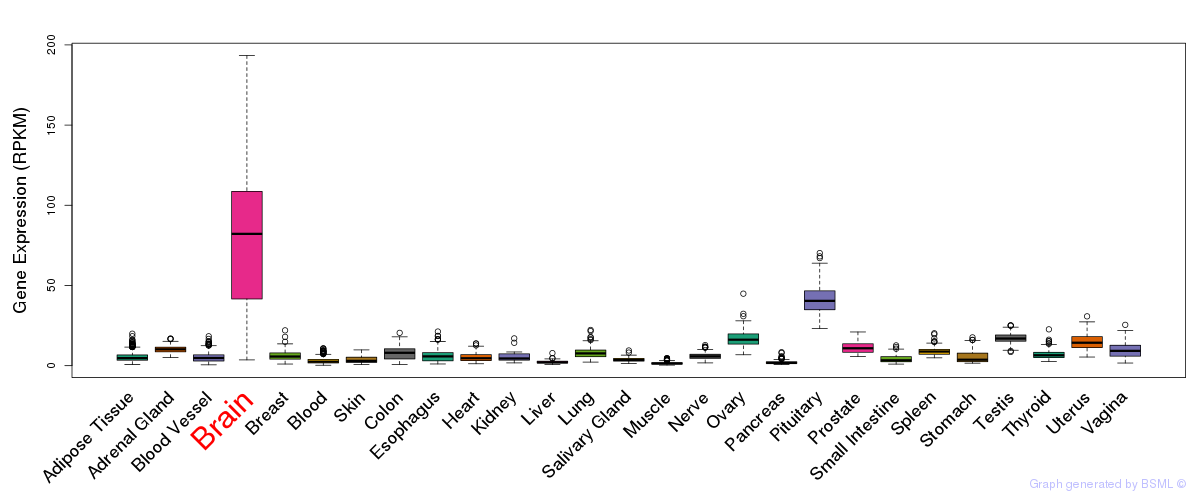
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0008022 | protein C-terminus binding | IPI | 7477295 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016188 | synaptic vesicle maturation | IEA | Synap (GO term level: 8) | - |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 9853749 |
| GO:0048169 | regulation of long-term neuronal synaptic plasticity | IEA | neuron, Synap (GO term level: 10) | - |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 9286702 |
| GO:0006461 | protein complex assembly | TAS | 7477295 | |
| GO:0007165 | signal transduction | TAS | 10364559 | |
| GO:0007612 | learning | TAS | 9853749 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0014069 | postsynaptic density | IEA | Synap (GO term level: 10) | - |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0045211 | postsynaptic membrane | IDA | Synap, Neurotransmitter (GO term level: 5) | 12151521 |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - | |
| GO:0031234 | extrinsic to internal side of plasma membrane | IEA | - | |
| GO:0030863 | cortical cytoskeleton | IDA | 12151521 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACCN3 | ASIC3 | DRASIC | SLNAC1 | TNaC1 | amiloride-sensitive cation channel 3 | ASIC3 interacts with DLG4 (PSD-95). This interaction was modeled on a demonstrated interaction between mouse or rat ASIC3 and rat DLG4 (PSD-95). | BIND | 15317815 |
| ACCN3 | ASIC3 | DRASIC | SLNAC1 | TNaC1 | amiloride-sensitive cation channel 3 | - | HPRD,BioGRID | 15317815 |
| ACTN2 | CMD1AA | actinin, alpha 2 | - | HPRD,BioGRID | 12435606 |
| ADAM22 | MDC2 | MGC149832 | ADAM metallopeptidase domain 22 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 16990550 |
| ADRB1 | ADRB1R | B1AR | BETA1AR | RHR | adrenergic, beta-1-, receptor | - | HPRD,BioGRID | 10995758 |
| AKAP5 | AKAP75 | AKAP79 | H21 | A kinase (PRKA) anchor protein 5 | - | HPRD | 10939335 |
| ATP2B2 | PMCA2 | PMCA2a | PMCA2i | ATPase, Ca++ transporting, plasma membrane 2 | - | HPRD | 11274188 |
| ATP2B2 | PMCA2 | PMCA2a | PMCA2i | ATPase, Ca++ transporting, plasma membrane 2 | PMCA2b interacts with SAP90. This interaction was modeled on a demonstrated interaction between human PMCA2b and rat SAP90. | BIND | 11274188 |
| ATP2B4 | ATP2B2 | DKFZp686G08106 | DKFZp686M088 | MXRA1 | PMCA4 | PMCA4b | PMCA4x | ATPase, Ca++ transporting, plasma membrane 4 | PMCA4b interacts with PSD-95. | BIND | 9430700 |
| ATP2B4 | ATP2B2 | DKFZp686G08106 | DKFZp686M088 | MXRA1 | PMCA4 | PMCA4b | PMCA4x | ATPase, Ca++ transporting, plasma membrane 4 | - | HPRD | 11274188 |
| BAI1 | FLJ41988 | brain-specific angiogenesis inhibitor 1 | - | HPRD,BioGRID | 11937501 |
| BEGAIN | KIAA1446 | brain-enriched guanylate kinase-associated homolog (rat) | - | HPRD | 9756850 |
| CACNG2 | MGC138502 | MGC138504 | calcium channel, voltage-dependent, gamma subunit 2 | - | HPRD,BioGRID | 11805122 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | - | HPRD,BioGRID | 12151521 |
| CD46 | MCP | MGC26544 | MIC10 | TLX | TRA2.10 | CD46 molecule, complement regulatory protein | - | HPRD,BioGRID | 11714708 |
| CIT | CRIK | KIAA0949 | STK21 | citron (rho-interacting, serine/threonine kinase 21) | in vivo Two-hybrid | BioGRID | 9870942 |
| CRIPT | HSPC139 | cysteine-rich PDZ-binding protein | - | HPRD,BioGRID | 9581762 |
| DAP | MGC99796 | death-associated protein | - | HPRD | 9286858 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | Affinity Capture-Western | BioGRID | 9286858 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | - | HPRD,BioGRID | 9182804 |
| DLG3 | KIAA1232 | MRX | MRX90 | NE-Dlg | NEDLG | SAP102 | discs, large homolog 3 (neuroendocrine-dlg, Drosophila) | - | HPRD,BioGRID | 10026200 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | Affinity Capture-Western | BioGRID | 9182804 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | Affinity Capture-Western Co-fractionation Far Western Reconstituted Complex Two-hybrid | BioGRID | 9024696 |9115257 |9286858 |10527873 |10844022 |11060025 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | - | HPRD | 9024696 |9286858 |
| DLGAP1 | DAP-1 | DAP-1-ALPHA | DAP-1-BETA | GKAP | MGC88156 | SAPAP1 | hGKAP | discs, large (Drosophila) homolog-associated protein 1 | PSD-95 interacts with DAP-1alpha. | BIND | 9286858 |
| DLGAP2 | DAP2 | SAPAP2 | discs, large (Drosophila) homolog-associated protein 2 | - | HPRD | 9286858 |
| DLGAP2 | DAP2 | SAPAP2 | discs, large (Drosophila) homolog-associated protein 2 | Reconstituted Complex Two-hybrid | BioGRID | 9115257 |
| DLGAP3 | DAP3 | SAPAP3 | discs, large (Drosophila) homolog-associated protein 3 | Two-hybrid | BioGRID | 9115257 |
| DLGAP4 | DAP4 | KIAA0964 | MGC131862 | RP5-977B1.6 | SAPAP4 | discs, large (Drosophila) homolog-associated protein 4 | Two-hybrid | BioGRID | 9115257 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10844022 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | - | HPRD | 14760703 |
| DYNLL2 | DNCL1B | Dlc2 | MGC17810 | dynein, light chain, LC8-type 2 | - | HPRD,BioGRID | 10844022 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD,BioGRID | 10839362 |
| ERBB2IP | ERBIN | LAP2 | erbb2 interacting protein | - | HPRD,BioGRID | 11279080 |
| ERBB4 | HER4 | MGC138404 | p180erbB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | - | HPRD,BioGRID | 10725395 |
| ERBB4 | HER4 | MGC138404 | p180erbB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | ErbB-4 interacts with PSD-95. | BIND | 10725395 |
| EXOC4 | MGC27170 | REC8 | SEC8 | SEC8L1 | Sec8p | exocyst complex component 4 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12675619 |12738960 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 12419528 |
| FZD1 | DKFZp564G072 | frizzled homolog 1 (Drosophila) | - | HPRD,BioGRID | 12067714 |
| FZD2 | - | frizzled homolog 2 (Drosophila) | Two-hybrid | BioGRID | 12067714 |
| FZD4 | CD344 | EVR1 | FEVR | FZD4S | Fz-4 | FzE4 | GPCR | MGC34390 | frizzled homolog 4 (Drosophila) | - | HPRD,BioGRID | 12067714 |
| FZD7 | FzE3 | frizzled homolog 7 (Drosophila) | Affinity Capture-Western Two-hybrid | BioGRID | 12067714 |
| GDA | CYPIN | GUANASE | KIAA1258 | MGC9982 | NEDASIN | guanine deaminase | - | HPRD | 10595517 |
| GRIA2 | GLUR2 | GLURB | GluR-K2 | HBGR2 | glutamate receptor, ionotropic, AMPA 2 | Affinity Capture-MS | BioGRID | 16990550 |
| GRIA3 | GLUR-C | GLUR-K3 | GLUR3 | GLURC | MRX94 | glutamate receptor, ionotrophic, AMPA 3 | Affinity Capture-MS | BioGRID | 16990550 |
| GRIK1 | EAA3 | EEA3 | GLR5 | GLUR5 | glutamate receptor, ionotropic, kainate 1 | - | HPRD,BioGRID | 12597860 |
| GRIK2 | EAA4 | GLR6 | GLUK6 | GLUR6 | MGC74427 | MRT6 | glutamate receptor, ionotropic, kainate 2 | - | HPRD | 9808460 |11276111 |12597860 |
| GRIK2 | EAA4 | GLR6 | GLUK6 | GLUR6 | MGC74427 | MRT6 | glutamate receptor, ionotropic, kainate 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 9808460 |11279111 |12597860 |
| GRIK5 | EAA2 | GRIK2 | KA2 | glutamate receptor, ionotropic, kainate 5 | - | HPRD,BioGRID | 9808460 |11279111 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 9009191 |9502803 |11506858 |12068077 |14732708 |15030493 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | - | HPRD | 7569905 |11937501 |12419528 |
| GRIN2A | NMDAR2A | NR2A | glutamate receptor, ionotropic, N-methyl D-aspartate 2A | Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 7569905 |9278515 |9286858 |10648730 |11937501 |12419528 |
| GRIN2B | MGC142178 | MGC142180 | NMDAR2B | NR2B | hNR3 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 7569905 |9278515 |9581762 |11937501 |11997254 |12738960 |
| GRIN2B | MGC142178 | MGC142180 | NMDAR2B | NR2B | hNR3 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | - | HPRD | 7569905 |9581762|11937501 |
| GRIN2C | NMDAR2C | NR2C | glutamate receptor, ionotropic, N-methyl D-aspartate 2C | - | HPRD,BioGRID | 7569905 |11937501 |
| GRIN2D | EB11 | NMDAR2D | glutamate receptor, ionotropic, N-methyl D-aspartate 2D | - | HPRD,BioGRID | 7569905 |
| GRIN3A | FLJ45414 | NMDAR-L | NR3A | glutamate receptor, ionotropic, N-methyl-D-aspartate 3A | in vitro in vivo Two-hybrid | BioGRID | 7569905 |
| GRIN3B | NR3B | glutamate receptor, ionotropic, N-methyl-D-aspartate 3B | in vitro in vivo Two-hybrid | BioGRID | 7569905 |
| GUCY1A2 | GC-SA2 | GUC1A2 | guanylate cyclase 1, soluble, alpha 2 | - | HPRD,BioGRID | 11572861 |
| HGS | HRS | ZFYVE8 | hepatocyte growth factor-regulated tyrosine kinase substrate | - | HPRD,BioGRID | 12151521 |
| HTR2A | 5-HT2A | HTR2 | 5-hydroxytryptamine (serotonin) receptor 2A | - | HPRD,BioGRID | 12682061 |
| HTR2C | 5-HT2C | 5-HTR2C | HTR1C | 5-hydroxytryptamine (serotonin) receptor 2C | - | HPRD,BioGRID | 12006486 |
| HTT | HD | IT15 | huntingtin | Huntingtin interacts with PSD-95. | BIND | 11319238 |
| IL13RA1 | CD213A1 | IL-13Ra | NR4 | interleukin 13 receptor, alpha 1 | in vitro in vivo Two-hybrid | BioGRID | 7569905 |
| KCNA1 | AEMK | EA1 | HBK1 | HUK1 | KV1.1 | MBK1 | MGC126782 | MGC138385 | MK1 | RBK1 | potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) | Reconstituted Complex Two-hybrid | BioGRID | 12435606 |
| KCNA2 | HBK5 | HK4 | HUKIV | KV1.2 | MGC50217 | MK2 | NGK1 | RBK2 | potassium voltage-gated channel, shaker-related subfamily, member 2 | - | HPRD,BioGRID | 8938729 |12435606 |
| KCNA4 | HBK4 | HK1 | HPCN2 | HUKII | KCNA4L | KCNA8 | KV1.4 | PCN2 | potassium voltage-gated channel, shaker-related subfamily, member 4 | - | HPRD | 8938729 |9581762 |12435606 |
| KCNA4 | HBK4 | HK1 | HPCN2 | HUKII | KCNA4L | KCNA8 | KV1.4 | PCN2 | potassium voltage-gated channel, shaker-related subfamily, member 4 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 8938729 |9581762 |11997254 |12435606 |
| KCNA5 | ATFB7 | HCK1 | HK2 | HPCN1 | KV1.5 | MGC117058 | MGC117059 | PCN1 | potassium voltage-gated channel, shaker-related subfamily, member 5 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12435606 |12860415 |
| KCND2 | KIAA1044 | KV4.2 | MGC119702 | MGC119703 | RK5 | potassium voltage-gated channel, Shal-related subfamily, member 2 | Two-hybrid | BioGRID | 12435606 |
| KCNJ10 | BIRK-10 | KCNJ13-PEN | KIR1.2 | KIR4.1 | potassium inwardly-rectifying channel, subfamily J, member 10 | - | HPRD | 9148889 |
| KCNJ12 | FLJ14167 | IRK2 | KCNJN1 | Kir2.2 | Kir2.2v | hIRK | hIRK1 | hkir2.2x | kcnj12x | potassium inwardly-rectifying channel, subfamily J, member 12 | - | HPRD,BioGRID | 15024025 |
| KCNJ2 | HHBIRK1 | HHIRK1 | IRK1 | KIR2.1 | LQT7 | SQT3 | potassium inwardly-rectifying channel, subfamily J, member 2 | - | HPRD,BioGRID | 10627592 |
| KCNJ4 | HIR | HIRK2 | HRK1 | IRK3 | Kir2.3 | MGC142066 | MGC142068 | potassium inwardly-rectifying channel, subfamily J, member 4 | Affinity Capture-Western Two-hybrid | BioGRID | 10627592 |11997254 |14960569 |
| KIF13B | GAKIN | KIAA0639 | kinesin family member 13B | - | HPRD,BioGRID | 10859302 |
| KIF1B | CMT2 | CMT2A | CMT2A1 | FLJ23699 | HMSNII | KIAA0591 | KIAA1448 | KLP | MGC134844 | kinesin family member 1B | - | HPRD | 12097473 |
| LGI1 | EPITEMPIN | EPT | ETL1 | IB1099 | leucine-rich, glioma inactivated 1 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 16990550 |
| LIN7A | LIN-7A | LIN7 | MALS-1 | MGC148143 | TIP-33 | VELI1 | lin-7 homolog A (C. elegans) | - | HPRD,BioGRID | 10341223 |
| LIN7B | LIN-7B | MALS-2 | MALS2 | VELI2 | lin-7 homolog B (C. elegans) | in vivo | BioGRID | 10341223 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD,BioGRID | 10827173 |
| LRP2 | DBS | gp330 | low density lipoprotein-related protein 2 | - | HPRD,BioGRID | 12713445 |
| LRP8 | APOER2 | HSZ75190 | MCI1 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor | Two-hybrid | BioGRID | 10827173 |
| LRRC1 | FLJ10775 | FLJ11834 | LANO | dJ523E19.1 | leucine rich repeat containing 1 | - | HPRD,BioGRID | 11440998 |
| LRRC7 | DKFZp686I1147 | KIAA1365 | MGC144918 | leucine rich repeat containing 7 | Affinity Capture-Western | BioGRID | 12390249 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD | 9892651 |
| MAP1A | FLJ77111 | MAP1L | MTAP1A | microtubule-associated protein 1A | - | HPRD,BioGRID | 9786987 |11925566 |
| MAP3K10 | MLK2 | MST | mitogen-activated protein kinase kinase kinase 10 | - | HPRD,BioGRID | 11152698 |
| MAP3K11 | MGC17114 | MLK-3 | MLK3 | PTK1 | SPRK | mitogen-activated protein kinase kinase kinase 11 | Affinity Capture-Western | BioGRID | 11152698 |
| NDOR1 | MGC138148 | NR1 | bA350O14.9 | NADPH dependent diflavin oxidoreductase 1 | - | HPRD,BioGRID | 7569905 |
| NLGN1 | KIAA1070 | MGC45115 | neuroligin 1 | Reconstituted Complex Two-hybrid | BioGRID | 9278515 |
| NLGN2 | KIAA1366 | neuroligin 2 | Two-hybrid | BioGRID | 9278515 |
| NLGN3 | ASPGX1 | AUTSX1 | HNL3 | KIAA1480 | neuroligin 3 | - | HPRD,BioGRID | 9278515 |
| NLGN4X | ASPGX2 | AUTSX2 | HLNX | HNLX | KIAA1260 | MGC22376 | NLGN | NLGN4 | neuroligin 4, X-linked | - | HPRD,BioGRID | 11368788 |
| NOS1 | IHPS1 | NOS | nNOS | nitric oxide synthase 1 (neuronal) | - | HPRD,BioGRID | 8625413 |
| NRG1 | ARIA | GGF | GGF2 | HGL | HRG | HRG1 | HRGA | NDF | SMDF | neuregulin 1 | Nrg-1 interacts with PSD-95 promoter. This interaction was modeled on a demonstrated interaction between Nrg-1 from an unspecified species and human PSD-95 promoter. | BIND | 15494726 |
| PARK2 | AR-JP | LPRS2 | PDJ | PRKN | Parkinson disease (autosomal recessive, juvenile) 2, parkin | Affinity Capture-Western | BioGRID | 11679592 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | - | HPRD,BioGRID | 11937501 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 12576483 |
| PTPRG | HPTPG | PTPG | R-PTP-GAMMA | RPTPG | protein tyrosine phosphatase, receptor type, G | - | HPRD,BioGRID | 10521598 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | - | HPRD | 12531901 |
| SEMA4C | FLJ20369 | KIAA1739 | M-SEMA-F | MGC126382 | MGC126383 | SEMACL1 | SEMAF | SEMAI | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C | - | HPRD,BioGRID | 11134026 |
| SEMA4F | M-SEMA | PRO2353 | SEMAM | SEMAW | m-Sema-M | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F | - | HPRD,BioGRID | 11483650 |
| SHANK1 | SPANK-1 | SSTRIP | synamon | SH3 and multiple ankyrin repeat domains 1 | - | HPRD,BioGRID | 10433268 |
| SHANK2 | CORTBP1 | CTTNBP1 | ProSAP1 | SHANK | SPANK-3 | SH3 and multiple ankyrin repeat domains 2 | - | HPRD,BioGRID | 10527873 |
| SIPA1L1 | DKFZp686G1344 | E6TP1 | KIAA0440 | signal-induced proliferation-associated 1 like 1 | - | HPRD,BioGRID | 11502259 |
| SYNGAP1 | DKFZp761G1421 | KIAA1938 | RASA1 | RASA5 | SYNGAP | synaptic Ras GTPase activating protein 1 homolog (rat) | Two-hybrid | BioGRID | 9581761 |
| TANC1 | KIAA1728 | ROLSB | TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | - | HPRD | 15673434 |
| YES1 | HsT441 | P61-YES | Yes | c-yes | v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 | - | HPRD | 9892651 |
| ZDHHC17 | HIP14 | HIP3 | HSPC294 | HYPH | KIAA0946 | zinc finger, DHHC-type containing 17 | HIP14 catalyzes palmitoylation of PSD-95. This interaction was modeled on a demonstrated interaction between human HIP14 and PSD-95 from an unspecified species. | BIND | 15603740 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NOS1 PATHWAY | 24 | 21 | All SZGR 2.0 genes in this pathway |
| ST GA12 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| ST GA13 PATHWAY | 37 | 32 | All SZGR 2.0 genes in this pathway |
| ST MYOCYTE AD PATHWAY | 27 | 25 | All SZGR 2.0 genes in this pathway |
| PID ERBB4 PATHWAY | 38 | 32 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| WATTEL AUTONOMOUS THYROID ADENOMA UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM2 | 153 | 102 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| RAMPON ENRICHED LEARNING ENVIRONMENT LATE UP | 22 | 16 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| LEIN LOCALIZED TO DISTAL AND PROXIMAL DENDRITES | 17 | 12 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR DN | 277 | 166 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 36HR | 152 | 88 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| ABDELMOHSEN ELAVL4 TARGETS | 16 | 13 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 339 | 346 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-103/107 | 747 | 753 | m8 | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-122 | 154 | 160 | m8 | hsa-miR-122a | UGGAGUGUGACAAUGGUGUUUGU |
| miR-149 | 728 | 734 | m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.