Gene Page: DRD1
Summary ?
| GeneID | 1812 |
| Symbol | DRD1 |
| Synonyms | DADR|DRD1A |
| Description | dopamine receptor D1 |
| Reference | MIM:126449|HGNC:HGNC:3020|Ensembl:ENSG00000184845|HPRD:00538|Vega:OTTHUMG00000130557 |
| Gene type | protein-coding |
| Map location | 5q35.1 |
| Pascal p-value | 0.107 |
| Fetal beta | -0.507 |
| eGene | Meta |
| Support | DOPAMINE GPCR SIGNALLING Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.0276 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 9 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 31 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
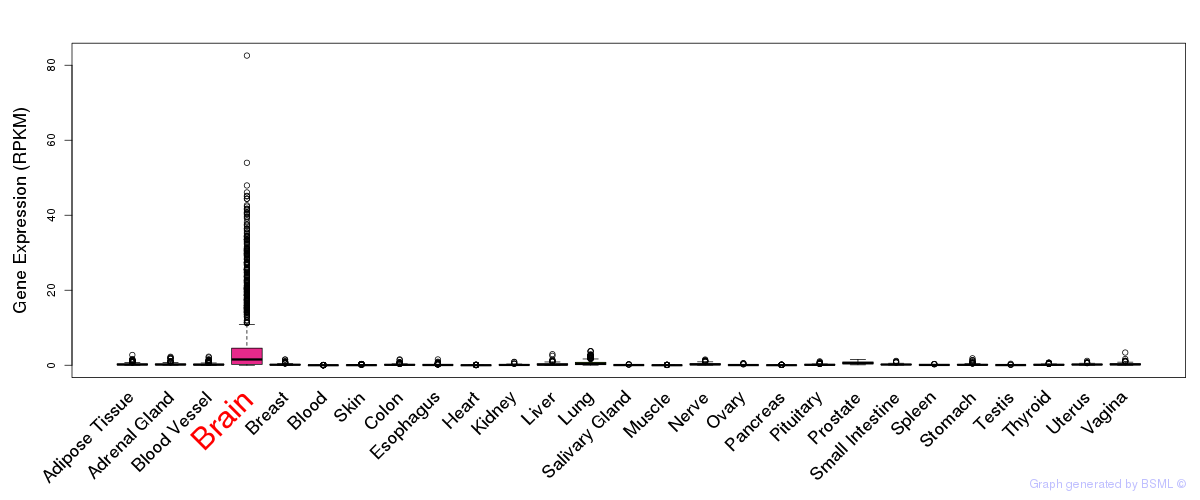
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf42 | 0.78 | 0.80 |
| AADAT | 0.77 | 0.76 |
| TCTN2 | 0.77 | 0.74 |
| KCTD3 | 0.76 | 0.80 |
| PROM1 | 0.75 | 0.75 |
| ZC3H7A | 0.74 | 0.81 |
| REV3L | 0.74 | 0.82 |
| MGEA5 | 0.74 | 0.78 |
| TOR1B | 0.74 | 0.76 |
| BIVM | 0.72 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| REEP6 | -0.54 | -0.49 |
| AF347015.31 | -0.54 | -0.65 |
| MT-CO2 | -0.53 | -0.65 |
| AF347015.27 | -0.53 | -0.62 |
| TSC22D4 | -0.52 | -0.56 |
| IFI27 | -0.51 | -0.62 |
| CA4 | -0.51 | -0.54 |
| FXYD1 | -0.51 | -0.59 |
| AF347015.33 | -0.51 | -0.60 |
| AIFM3 | -0.51 | -0.51 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001590 | dopamine D1 receptor activity | IEA | dopamine (GO term level: 10) | - |
| GO:0004872 | receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007416 | synaptogenesis | ISS | Synap (GO term level: 6) | - |
| GO:0060158 | dopamine receptor, phospholipase C activating pathway | IGI | dopamine (GO term level: 10) | 17194762 |
| GO:0051584 | regulation of dopamine uptake | IC | Synap, Neurotransmitter, dopamine (GO term level: 11) | 8301582 |
| GO:0051968 | positive regulation of synaptic transmission, glutamatergic | ISS | neuron, glutamate, Synap, Neurotransmitter (GO term level: 10) | - |
| GO:0007191 | dopamine receptor, adenylate cyclase activating pathway | IEA | dopamine (GO term level: 12) | - |
| GO:0019226 | transmission of nerve impulse | IEA | neuron (GO term level: 5) | - |
| GO:0021853 | cerebral cortex GABAergic interneuron migration | IEA | neuron, GABA (GO term level: 14) | - |
| GO:0042053 | regulation of dopamine metabolic process | IEA | dopamine (GO term level: 9) | - |
| GO:0001975 | response to amphetamine | IEA | - | |
| GO:0007187 | G-protein signaling, coupled to cyclic nucleotide second messenger | IDA | 1282671 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008542 | visual learning | IEA | - | |
| GO:0010580 | activation of adenylate cyclase activity involved in G-protein signaling | IDA | 2144334 | |
| GO:0007631 | feeding behavior | IEA | - | |
| GO:0007628 | adult walking behavior | IEA | - | |
| GO:0007626 | locomotory behavior | IEA | - | |
| GO:0007625 | grooming behavior | IEA | - | |
| GO:0007617 | mating behavior | IEA | - | |
| GO:0042493 | response to drug | IEA | - | |
| GO:0030432 | peristalsis | IEA | - | |
| GO:0051281 | positive regulation of release of sequestered calcium ion into cytosol | IDA | 1282671 |7838121 | |
| GO:0030819 | positive regulation of cAMP biosynthetic process | IDA | 1282671 |7838121 | |
| GO:0030335 | positive regulation of cell migration | IEA | - | |
| GO:0043268 | positive regulation of potassium ion transport | ISS | - | |
| GO:0051482 | elevation of cytosolic calcium ion concentration during G-protein signaling, coupled to IP3 second messenger (phospholipase C activating) | IDA | 1282671 | |
| GO:0060134 | prepulse inhibition | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005624 | membrane fraction | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 11500503 | |
| GO:0005887 | integral to plasma membrane | IC | 2144334 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| KEGG GAP JUNCTION | 90 | 68 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CK1 PATHWAY | 17 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINE LIGAND BINDING RECEPTORS | 38 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER DN | 28 | 26 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| BRIDEAU IMPRINTED GENES | 63 | 47 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-15/16/195/424/497 | 117 | 123 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-203.1 | 1060 | 1066 | 1A | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-216 | 693 | 699 | m8 | hsa-miR-216 | UAAUCUCAGCUGGCAACUGUG |
| miR-29 | 380 | 386 | 1A | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-496 | 1028 | 1034 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-500 | 423 | 429 | m8 | hsa-miR-500 | AUGCACCUGGGCAAGGAUUCUG |
| miR-539 | 1055 | 1061 | 1A | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 654 | 660 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.