Gene Page: DVL2
Summary ?
| GeneID | 1856 |
| Symbol | DVL2 |
| Synonyms | - |
| Description | dishevelled segment polarity protein 2 |
| Reference | MIM:602151|HGNC:HGNC:3086|Ensembl:ENSG00000004975|HPRD:03690|Vega:OTTHUMG00000102155 |
| Gene type | protein-coding |
| Map location | 17p13.1 |
| Pascal p-value | 1.528E-4 |
| Sherlock p-value | 0.274 |
| Fetal beta | 0.361 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1522 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13248847 | 17 | 7137857 | DVL2 | 3.88E-5 | -0.212 | 0.02 | DMG:Wockner_2014 |
| cg11181795 | 17 | 7219971 | DVL2 | 3.03E-4 | -4.25 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
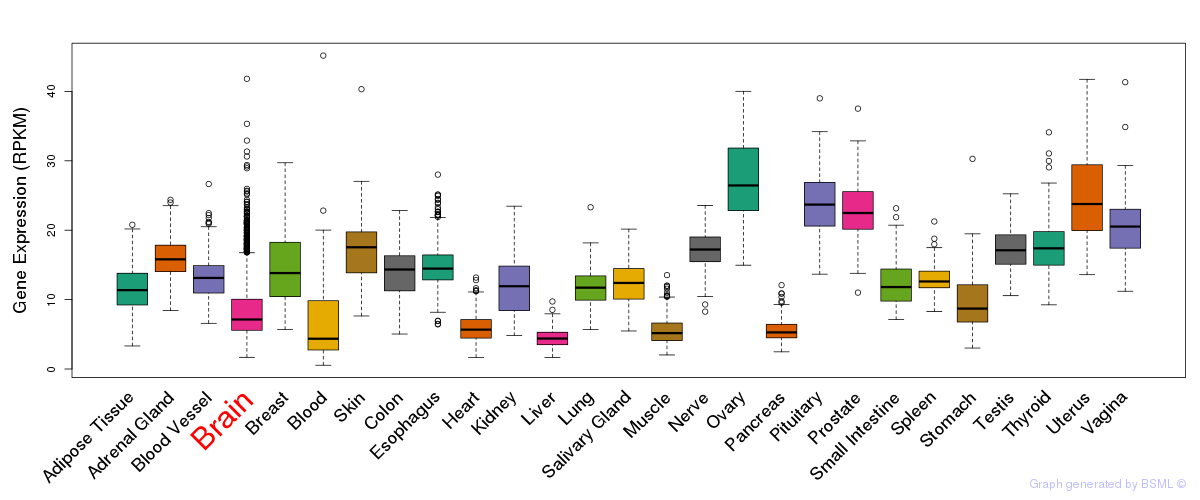
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | TAS | 8887313 | |
| GO:0042802 | identical protein binding | IPI | 16189514 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001843 | neural tube closure | IEA | - | |
| GO:0016055 | Wnt receptor signaling pathway | IEA | - | |
| GO:0007379 | segment specification | IEA | - | |
| GO:0007242 | intracellular signaling cascade | IEA | - | |
| GO:0007507 | heart development | IEA | - | |
| GO:0007447 | imaginal disc pattern formation | TAS | 8887313 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | TAS | 9192851 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AP1M1 | AP47 | CLAPM2 | CLTNM | MU-1A | adaptor-related protein complex 1, mu 1 subunit | Two-hybrid | BioGRID | 16189514 |
| AP2M1 | AP50 | CLAPM1 | mu2 | adaptor-related protein complex 2, mu 1 subunit | Two-hybrid | BioGRID | 16189514 |
| ARR3 | ARRX | arrestin 3, retinal (X-arrestin) | Two-hybrid | BioGRID | 16189514 |
| ARRB1 | ARB1 | ARR1 | arrestin, beta 1 | - | HPRD,BioGRID | 11742073 |
| AXIN1 | AXIN | MGC52315 | axin 1 | - | HPRD,BioGRID | 10829020 |
| BAG3 | BAG-3 | BIS | CAIR-1 | MGC104307 | BCL2-associated athanogene 3 | Two-hybrid | BioGRID | 16189514 |
| BAHD1 | KIAA0945 | bromo adjacent homology domain containing 1 | Two-hybrid | BioGRID | 16189514 |
| C16orf48 | DAKV6410 | DKFZp434A1319 | chromosome 16 open reading frame 48 | Two-hybrid | BioGRID | 16189514 |
| C9orf100 | FLJ14642 | MGC44886 | RP11-331F9.7 | chromosome 9 open reading frame 100 | Two-hybrid | BioGRID | 16189514 |
| CCDC33 | FLJ23168 | FLJ32855 | MGC34145 | coiled-coil domain containing 33 | Two-hybrid | BioGRID | 16189514 |
| CSNK1E | HCKIE | MGC10398 | casein kinase 1, epsilon | Two-hybrid | BioGRID | 16189514 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | Affinity Capture-Western | BioGRID | 10806215 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | Affinity Capture-Western | BioGRID | 10806215 |
| DAB2 | DOC-2 | DOC2 | FLJ26626 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | Affinity Capture-Western Reconstituted Complex | BioGRID | 12805222 |
| DDI1 | FLJ36017 | DDI1, DNA-damage inducible 1, homolog 1 (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| DPPA2 | PESCRG1 | developmental pluripotency associated 2 | Two-hybrid | BioGRID | 16189514 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| FAM105B | FLJ34884 | family with sequence similarity 105, member B | Two-hybrid | BioGRID | 16189514 |
| FLJ12529 | FLJ39024 | MGC9315 | pre-mRNA cleavage factor I, 59 kDa subunit | Two-hybrid | BioGRID | 16189514 |
| MAP3K1 | MAPKKK1 | MEKK | MEKK1 | mitogen-activated protein kinase kinase kinase 1 | Affinity Capture-Western | BioGRID | 10829020 |
| MCRS1 | ICP22BP | INO80Q | MCRS2 | MSP58 | P78 | microspherule protein 1 | Two-hybrid | BioGRID | 16189514 |
| PCBD1 | DCOH | PCBD | PCD | PHS | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha | Two-hybrid | BioGRID | 16189514 |
| POLI | RAD30B | RAD3OB | polymerase (DNA directed) iota | Two-hybrid | BioGRID | 16189514 |
| PRPF3 | HPRP3 | HPRP3P | PRP3 | Prp3p | RP18 | PRP3 pre-mRNA processing factor 3 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| RBPMS | HERMES | RNA binding protein with multiple splicing | Two-hybrid | BioGRID | 16189514 |
| RHOXF2 | PEPP-2 | PEPP2 | THG1 | Rhox homeobox family, member 2 | Two-hybrid | BioGRID | 16189514 |
| RNPS1 | E5.1 | MGC117332 | RNA binding protein S1, serine-rich domain | Two-hybrid | BioGRID | 16189514 |
| SNF8 | Dot3 | EAP30 | VPS22 | SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16189514 |
| SNIP1 | FLJ12553 | RP3-423B22.3 | dJ423B22.2 | Smad nuclear interacting protein 1 | Two-hybrid | BioGRID | 16189514 |
| SORBS3 | SCAM-1 | SCAM1 | SH3D4 | VINEXIN | sorbin and SH3 domain containing 3 | Two-hybrid | BioGRID | 16189514 |
| TCEB3B | ELOA2 | HsT832 | MGC119351 | TCEB3L | transcription elongation factor B polypeptide 3B (elongin A2) | Two-hybrid | BioGRID | 16189514 |
| THAP1 | FLJ10477 | MGC33014 | THAP domain containing, apoptosis associated protein 1 | Two-hybrid | BioGRID | 16189514 |
| TIFA | MGC20791 | T2BP | T6BP | TIFAA | TRAF-interacting protein with forkhead-associated domain | Two-hybrid | BioGRID | 16189514 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Two-hybrid | BioGRID | 16189514 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Two-hybrid | BioGRID | 16189514 |
| U2AF2 | U2AF65 | U2 small nuclear RNA auxiliary factor 2 | Two-hybrid | BioGRID | 16189514 |
| USP5 | ISOT | ubiquitin specific peptidase 5 (isopeptidase T) | Two-hybrid | BioGRID | 16189514 |
| VANGL1 | LPP2 | MGC5338 | STB2 | STBM2 | vang-like 1 (van gogh, Drosophila) | - | HPRD | 15456783 |
| ZBTB8 | BOZF1 | FLJ90065 | MGC17919 | zinc finger and BTB domain containing 8 | Two-hybrid | BioGRID | 16189514 |
| ZNF165 | LD65 | ZSCAN7 | zinc finger protein 165 | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| ZNF250 | FLJ57354 | MGC111123 | MGC9718 | ZFP647 | ZNF647 | zinc finger protein 250 | Two-hybrid | BioGRID | 16189514 |
| ZNF263 | FPM315 | ZKSCAN12 | zinc finger protein 263 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| KEGG NOTCH SIGNALING PATHWAY | 47 | 35 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG BASAL CELL CARCINOMA | 55 | 44 | All SZGR 2.0 genes in this pathway |
| WNT SIGNALING | 89 | 71 | All SZGR 2.0 genes in this pathway |
| PID WNT NONCANONICAL PATHWAY | 32 | 26 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN DEG PATHWAY | 18 | 17 | All SZGR 2.0 genes in this pathway |
| PID WNT CANONICAL PATHWAY | 20 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY HIPPO | 22 | 15 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 1 | 13 | 11 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIN ACTION DIRECT VS INDIRECT 4HR | 37 | 22 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 126 | 133 | 1A,m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-138 | 138 | 145 | 1A,m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-27 | 127 | 133 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-495 | 460 | 466 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.