Gene Page: FAH
Summary ?
| GeneID | 2184 |
| Symbol | FAH |
| Synonyms | - |
| Description | fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| Reference | MIM:613871|HGNC:HGNC:3579|Ensembl:ENSG00000103876|HPRD:02040|Vega:OTTHUMG00000144187 |
| Gene type | protein-coding |
| Map location | 15q25.1 |
| Pascal p-value | 0.01 |
| Fetal beta | -0.794 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg07699440 | 15 | 80445357 | FAH | -0.021 | 0.42 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs34053230 | 15 | 80448562 | FAH | ENSG00000103876.7 | 1.702E-6 | 0 | 3730 | gtex_brain_ba24 |
| rs35271165 | 15 | 80448626 | FAH | ENSG00000103876.7 | 2.355E-10 | 0 | 3794 | gtex_brain_ba24 |
| rs71397596 | 15 | 80449583 | FAH | ENSG00000103876.7 | 2.567E-9 | 0 | 4751 | gtex_brain_ba24 |
| rs35880073 | 15 | 80452013 | FAH | ENSG00000103876.7 | 2.485E-9 | 0 | 7181 | gtex_brain_ba24 |
| rs33929922 | 15 | 80452172 | FAH | ENSG00000103876.7 | 2.485E-9 | 0 | 7340 | gtex_brain_ba24 |
| rs2014531 | 15 | 80452943 | FAH | ENSG00000103876.7 | 2.485E-9 | 0 | 8111 | gtex_brain_ba24 |
| rs11856427 | 15 | 80453914 | FAH | ENSG00000103876.7 | 9.262E-8 | 0 | 9082 | gtex_brain_ba24 |
| rs11852901 | 15 | 80453971 | FAH | ENSG00000103876.7 | 2.513E-9 | 0 | 9139 | gtex_brain_ba24 |
| rs12907580 | 15 | 80455234 | FAH | ENSG00000103876.7 | 2.18E-10 | 0 | 10402 | gtex_brain_ba24 |
| rs34497292 | 15 | 80458548 | FAH | ENSG00000103876.7 | 2.254E-9 | 0 | 13716 | gtex_brain_ba24 |
| rs202244731 | 15 | 80458981 | FAH | ENSG00000103876.7 | 2.668E-8 | 0 | 14149 | gtex_brain_ba24 |
| rs6495476 | 15 | 80458982 | FAH | ENSG00000103876.7 | 2.668E-8 | 0 | 14150 | gtex_brain_ba24 |
| rs12910663 | 15 | 80467964 | FAH | ENSG00000103876.7 | 2.321E-9 | 0 | 23132 | gtex_brain_ba24 |
| rs12912992 | 15 | 80470426 | FAH | ENSG00000103876.7 | 2.375E-9 | 0 | 25594 | gtex_brain_ba24 |
| rs7168949 | 15 | 80470457 | FAH | ENSG00000103876.7 | 3.959E-9 | 0 | 25625 | gtex_brain_ba24 |
| rs58626166 | 15 | 80471518 | FAH | ENSG00000103876.7 | 1.155E-8 | 0 | 26686 | gtex_brain_ba24 |
| rs71397597 | 15 | 80473662 | FAH | ENSG00000103876.7 | 1.872E-10 | 0 | 28830 | gtex_brain_ba24 |
| rs12898575 | 15 | 80478262 | FAH | ENSG00000103876.7 | 3.425E-9 | 0 | 33430 | gtex_brain_ba24 |
| rs12898576 | 15 | 80478268 | FAH | ENSG00000103876.7 | 3.425E-9 | 0 | 33436 | gtex_brain_ba24 |
| rs200464452 | 15 | 80478599 | FAH | ENSG00000103876.7 | 2.617E-9 | 0 | 33767 | gtex_brain_ba24 |
| rs1049194 | 15 | 80478645 | FAH | ENSG00000103876.7 | 2.617E-9 | 0 | 33813 | gtex_brain_ba24 |
| rs79203348 | 15 | 80478835 | FAH | ENSG00000103876.7 | 1.815E-10 | 0 | 34003 | gtex_brain_ba24 |
| rs75212096 | 15 | 80478861 | FAH | ENSG00000103876.7 | 2.503E-9 | 0 | 34029 | gtex_brain_ba24 |
| rs34245126 | 15 | 80479116 | FAH | ENSG00000103876.7 | 2.392E-9 | 0 | 34284 | gtex_brain_ba24 |
| rs12904314 | 15 | 80479620 | FAH | ENSG00000103876.7 | 2.236E-9 | 0 | 34788 | gtex_brain_ba24 |
| rs12904656 | 15 | 80479659 | FAH | ENSG00000103876.7 | 2.233E-9 | 0 | 34827 | gtex_brain_ba24 |
| rs12591328 | 15 | 80479884 | FAH | ENSG00000103876.7 | 2.215E-9 | 0 | 35052 | gtex_brain_ba24 |
| rs8041036 | 15 | 80481049 | FAH | ENSG00000103876.7 | 4.387E-9 | 0 | 36217 | gtex_brain_ba24 |
| rs1437222 | 15 | 80486164 | FAH | ENSG00000103876.7 | 5.572E-8 | 0 | 41332 | gtex_brain_ba24 |
| rs1437221 | 15 | 80486402 | FAH | ENSG00000103876.7 | 6.532E-9 | 0 | 41570 | gtex_brain_ba24 |
| rs34392733 | 15 | 80486539 | FAH | ENSG00000103876.7 | 9.016E-9 | 0 | 41707 | gtex_brain_ba24 |
| rs34606389 | 15 | 80487481 | FAH | ENSG00000103876.7 | 4.387E-9 | 0 | 42649 | gtex_brain_ba24 |
| rs12593705 | 15 | 80487626 | FAH | ENSG00000103876.7 | 2.137E-9 | 0 | 42794 | gtex_brain_ba24 |
| rs3087815 | 15 | 80487900 | FAH | ENSG00000103876.7 | 2.301E-9 | 0 | 43068 | gtex_brain_ba24 |
| rs12708527 | 15 | 80488668 | FAH | ENSG00000103876.7 | 4.417E-9 | 0 | 43836 | gtex_brain_ba24 |
| rs16971828 | 15 | 80489013 | FAH | ENSG00000103876.7 | 2.154E-9 | 0 | 44181 | gtex_brain_ba24 |
| rs35376511 | 15 | 80489251 | FAH | ENSG00000103876.7 | 1.807E-10 | 0 | 44419 | gtex_brain_ba24 |
| rs7165803 | 15 | 80490624 | FAH | ENSG00000103876.7 | 3.085E-10 | 0 | 45792 | gtex_brain_ba24 |
| rs7166331 | 15 | 80490766 | FAH | ENSG00000103876.7 | 9.309E-11 | 0 | 45934 | gtex_brain_ba24 |
| rs12593336 | 15 | 80491786 | FAH | ENSG00000103876.7 | 1.405E-8 | 0 | 46954 | gtex_brain_ba24 |
| rs71397599 | 15 | 80499075 | FAH | ENSG00000103876.7 | 4.772E-8 | 0 | 54243 | gtex_brain_ba24 |
| rs71397600 | 15 | 80500149 | FAH | ENSG00000103876.7 | 4.772E-8 | 0 | 55317 | gtex_brain_ba24 |
| rs28692766 | 15 | 80500220 | FAH | ENSG00000103876.7 | 4.772E-8 | 0 | 55388 | gtex_brain_ba24 |
| rs34939304 | 15 | 80500367 | FAH | ENSG00000103876.7 | 4.772E-8 | 0 | 55535 | gtex_brain_ba24 |
| rs17219447 | 15 | 80501242 | FAH | ENSG00000103876.7 | 4.772E-8 | 0 | 56410 | gtex_brain_ba24 |
| rs12898706 | 15 | 80501402 | FAH | ENSG00000103876.7 | 4.772E-8 | 0 | 56570 | gtex_brain_ba24 |
| rs71397601 | 15 | 80507736 | FAH | ENSG00000103876.7 | 4.763E-8 | 0 | 62904 | gtex_brain_ba24 |
| rs12913384 | 15 | 80513833 | FAH | ENSG00000103876.7 | 1.421E-6 | 0 | 69001 | gtex_brain_ba24 |
| rs71397602 | 15 | 80514848 | FAH | ENSG00000103876.7 | 1.349E-6 | 0 | 70016 | gtex_brain_ba24 |
| rs12905402 | 15 | 80516383 | FAH | ENSG00000103876.7 | 1.349E-6 | 0 | 71551 | gtex_brain_ba24 |
| rs35971717 | 15 | 80517610 | FAH | ENSG00000103876.7 | 1.005E-6 | 0 | 72778 | gtex_brain_ba24 |
| rs199614927 | 15 | 80517770 | FAH | ENSG00000103876.7 | 1.302E-6 | 0 | 72938 | gtex_brain_ba24 |
| rs71399403 | 15 | 80517919 | FAH | ENSG00000103876.7 | 1.349E-6 | 0 | 73087 | gtex_brain_ba24 |
| rs35222819 | 15 | 80518506 | FAH | ENSG00000103876.7 | 4.851E-8 | 0 | 73674 | gtex_brain_ba24 |
| rs34765237 | 15 | 80520275 | FAH | ENSG00000103876.7 | 1.349E-6 | 0 | 75443 | gtex_brain_ba24 |
| rs80244168 | 15 | 80520961 | FAH | ENSG00000103876.7 | 1.349E-6 | 0 | 76129 | gtex_brain_ba24 |
| rs28621634 | 15 | 80521264 | FAH | ENSG00000103876.7 | 1.414E-6 | 0 | 76432 | gtex_brain_ba24 |
| rs71399405 | 15 | 80533155 | FAH | ENSG00000103876.7 | 1.348E-6 | 0 | 88323 | gtex_brain_ba24 |
| rs34424593 | 15 | 80539476 | FAH | ENSG00000103876.7 | 1.228E-6 | 0 | 94644 | gtex_brain_ba24 |
| rs12907596 | 15 | 80542852 | FAH | ENSG00000103876.7 | 8.184E-8 | 0 | 98020 | gtex_brain_ba24 |
| rs12903574 | 15 | 80551446 | FAH | ENSG00000103876.7 | 3.986E-9 | 0 | 106614 | gtex_brain_ba24 |
| rs12910345 | 15 | 80552343 | FAH | ENSG00000103876.7 | 3.986E-9 | 0 | 107511 | gtex_brain_ba24 |
| rs34457632 | 15 | 80555024 | FAH | ENSG00000103876.7 | 3.986E-9 | 0 | 110192 | gtex_brain_ba24 |
| rs1370272 | 15 | 80557770 | FAH | ENSG00000103876.7 | 3.986E-9 | 0 | 112938 | gtex_brain_ba24 |
| rs117618678 | 15 | 80572755 | FAH | ENSG00000103876.7 | 5.836E-9 | 0 | 127923 | gtex_brain_ba24 |
| rs7180132 | 15 | 80573960 | FAH | ENSG00000103876.7 | 6.484E-8 | 0 | 129128 | gtex_brain_ba24 |
| rs7174862 | 15 | 80588633 | FAH | ENSG00000103876.7 | 3.436E-7 | 0 | 143801 | gtex_brain_ba24 |
Section II. Transcriptome annotation
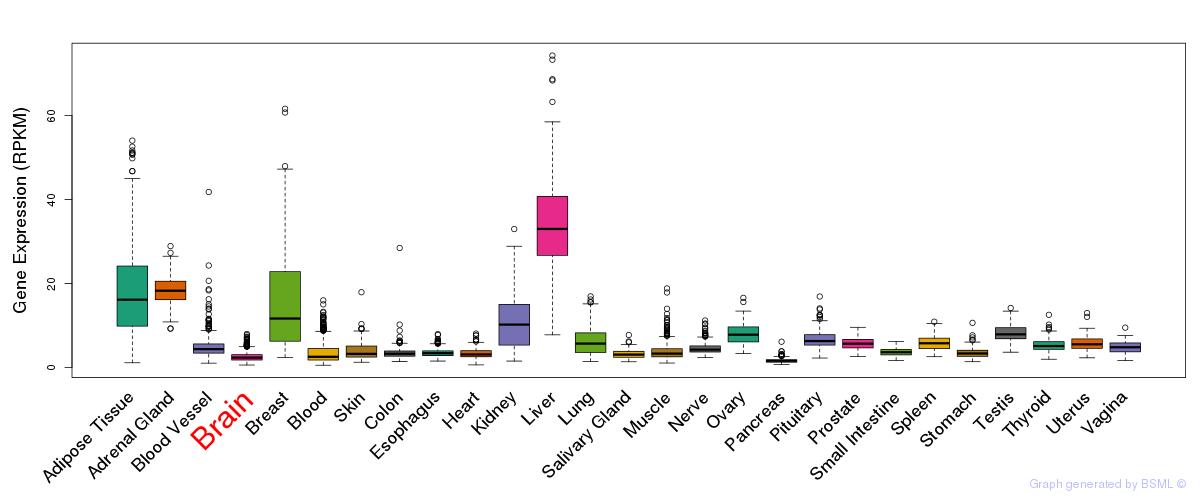
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LGR6 | 0.70 | 0.68 |
| C4orf19 | 0.70 | 0.70 |
| PPP1R3G | 0.68 | 0.68 |
| CHST7 | 0.68 | 0.57 |
| OAF | 0.67 | 0.66 |
| SFXN5 | 0.67 | 0.68 |
| PTH1R | 0.66 | 0.70 |
| ADORA2B | 0.66 | 0.62 |
| AC011427.1 | 0.65 | 0.61 |
| NEU4 | 0.65 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| POLR3C | -0.53 | -0.60 |
| ZNF7 | -0.52 | -0.60 |
| ZNF485 | -0.52 | -0.62 |
| DYNC1I2 | -0.52 | -0.63 |
| ZNF433 | -0.52 | -0.63 |
| TRA2A | -0.51 | -0.59 |
| EXOSC2 | -0.51 | -0.56 |
| ZBED5 | -0.51 | -0.62 |
| ZNF439 | -0.51 | -0.58 |
| ZNF226 | -0.51 | -0.64 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0004334 | fumarylacetoacetase activity | EXP | 8364576 | |
| GO:0004334 | fumarylacetoacetase activity | TAS | 1998338 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009072 | aromatic amino acid family metabolic process | IEA | - | |
| GO:0006527 | arginine catabolic process | IEA | - | |
| GO:0008152 | metabolic process | IEA | - | |
| GO:0006559 | L-phenylalanine catabolic process | IEA | - | |
| GO:0006572 | tyrosine catabolic process | TAS | 9305902 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 8364576 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TYROSINE METABOLISM | 42 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS DN | 73 | 51 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| HU ANGIOGENESIS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS BASAL | 330 | 217 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| WEI MIR34A TARGETS | 148 | 97 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGG VS IGA UP | 22 | 12 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS DN | 87 | 69 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 0HR | 63 | 48 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4 | 100 | 62 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G12 DN | 15 | 11 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT DN | 222 | 141 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| VANLOO SP3 TARGETS DN | 89 | 47 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 36HR | 152 | 88 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |