Gene Page: CABIN1
Summary ?
| GeneID | 23523 |
| Symbol | CABIN1 |
| Synonyms | CAIN|KB-318B8.7|PPP3IN |
| Description | calcineurin binding protein 1 |
| Reference | MIM:604251|HGNC:HGNC:24187|Ensembl:ENSG00000099991|HPRD:10369|Vega:OTTHUMG00000150797 |
| Gene type | protein-coding |
| Map location | 22q11.23 |
| Pascal p-value | 0.473 |
| Sherlock p-value | 0.909 |
| Fetal beta | -1.022 |
| eGene | Cortex |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 2.7575 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CABIN1 | chr22 | 24479335 | C | T | NM_001199281 NM_001201429 NM_012295 | p.968A>V p.918A>V p.968A>V | missense missense missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
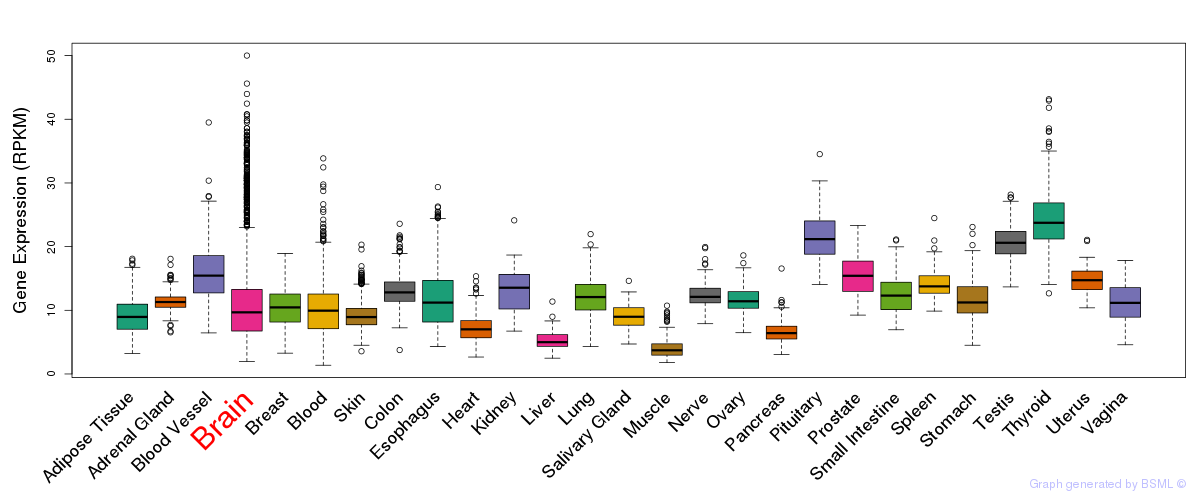
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PTGES2 | 0.92 | 0.94 |
| STUB1 | 0.90 | 0.94 |
| HAGH | 0.90 | 0.92 |
| ADAP1 | 0.90 | 0.90 |
| RAB4B | 0.89 | 0.89 |
| ABHD8 | 0.89 | 0.92 |
| RUNDC3A | 0.89 | 0.91 |
| PDXP | 0.88 | 0.89 |
| SNRPN | 0.88 | 0.91 |
| SH3BP1 | 0.87 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF5B | -0.53 | -0.61 |
| THOC2 | -0.45 | -0.43 |
| CCAR1 | -0.44 | -0.39 |
| AC005921.3 | -0.44 | -0.64 |
| ANKRD36B | -0.44 | -0.55 |
| SFRS18 | -0.43 | -0.55 |
| NSBP1 | -0.43 | -0.40 |
| ANKRD36 | -0.42 | -0.54 |
| ZNF560 | -0.42 | -0.44 |
| SFRS12 | -0.42 | -0.41 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005488 | binding | IEA | - | |
| GO:0004864 | protein phosphatase inhibitor activity | NAS | 9655484 | |
| GO:0008170 | N-methyltransferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006306 | DNA methylation | IEA | - | |
| GO:0007166 | cell surface receptor linked signal transduction | NAS | 9655484 | |
| GO:0016568 | chromatin modification | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | NAS | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AMPH | AMPH1 | amphiphysin | - | HPRD,BioGRID | 10931822 |
| AP1B1 | ADTB1 | AP105A | BAM22 | CLAPB2 | adaptor-related protein complex 1, beta 1 subunit | - | HPRD,BioGRID | 10931822 |
| CALM1 | CALML2 | CAMI | DD132 | PHKD | calmodulin 1 (phosphorylase kinase, delta) | CALM1 (CaM) interacts with CABIN1. This interaction was modelled on a demonstrated interaction between CaM from an unspecified species and human CABIN1. | BIND | 15902271 |
| CAMK4 | CaMK-GR | MGC36771 | calcium/calmodulin-dependent protein kinase IV | CAMK4 (CAMKIV) interacts with CABIN1. | BIND | 15902271 |
| DNM1 | DNM | dynamin 1 | - | HPRD,BioGRID | 10931822 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 10933397 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | - | HPRD,BioGRID | 10933397 |
| MEF2B | FLJ32599 | FLJ46391 | MGC189732 | MGC189763 | RSRFR2 | myocyte enhancer factor 2B | - | HPRD,BioGRID | 10531067 |12700764 |
| MEF2D | DKFZp686I1536 | myocyte enhancer factor 2D | Affinity Capture-Western Reconstituted Complex | BioGRID | 10531067 |10933397 |
| MEF2D | DKFZp686I1536 | myocyte enhancer factor 2D | MEF2D interacts with CABIN1. | BIND | 15902271 |
| PPP3CA | CALN | CALNA | CALNA1 | CCN1 | CNA1 | PPP2B | protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform | Two-hybrid | BioGRID | 9655484 |
| PPP3CA | CALN | CALNA | CALNA1 | CCN1 | CNA1 | PPP2B | protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform | - | HPRD | 11714752 |
| PPP3CB | CALNA2 | CALNB | protein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform | - | HPRD | 11714752 |
| PPP3CB | CALNA2 | CALNB | protein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform | - | HPRD | 9655484 |
| PPP3CC | CALNA3 | protein phosphatase 3 (formerly 2B), catalytic subunit, gamma isoform | - | HPRD | 11714752 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | - | HPRD,BioGRID | 10933397 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | YWHAQ (14-3-3-theta) interacts with phosphorylated CABIN1. This interaction was modelled on a demonstrated interaction between mouse 14-3-3-theta and human CABIN1. | BIND | 15902271 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA HDAC PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MEF2D PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| PID NFAT 3PATHWAY | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID TCR CALCIUM PATHWAY | 29 | 23 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 MCF10A | 43 | 26 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA UP | 75 | 43 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA DN | 80 | 53 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |