Gene Page: MLH3
Summary ?
| GeneID | 27030 |
| Symbol | MLH3 |
| Synonyms | HNPCC7 |
| Description | mutL homolog 3 |
| Reference | MIM:604395|HGNC:HGNC:7128|Ensembl:ENSG00000119684|HPRD:05094|Vega:OTTHUMG00000171766 |
| Gene type | protein-coding |
| Map location | 14q24.3 |
| Pascal p-value | 0.467 |
| Fetal beta | 0.747 |
| DMG | 1 (# studies) |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04798158 | 14 | 75179846 | MLH3 | 9.355E-4 | 6.262 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
General gene expression (GTEx)
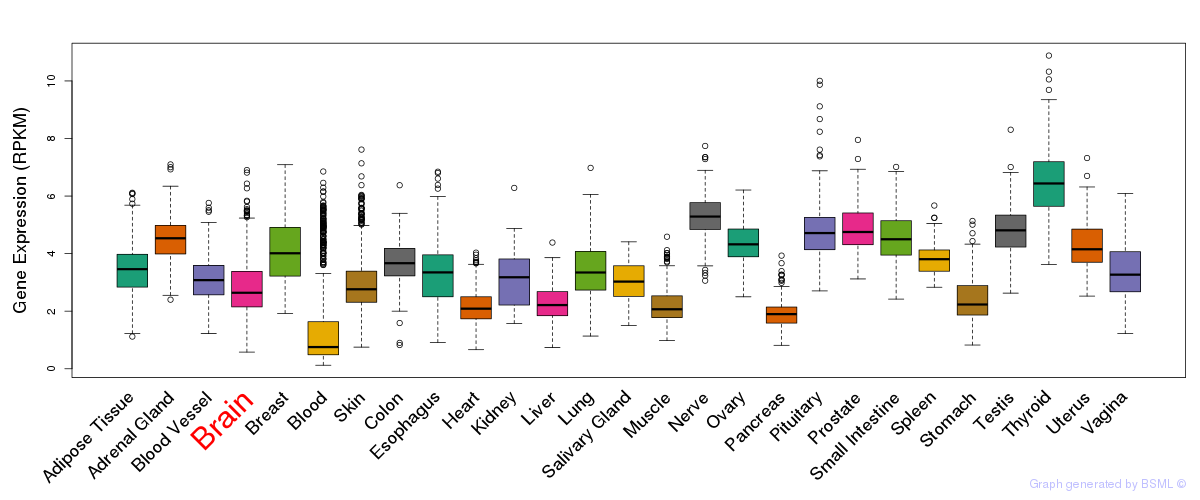
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MICAL1 | 0.87 | 0.84 |
| ZNF311 | 0.86 | 0.86 |
| ZNF133 | 0.86 | 0.83 |
| ADAMTS10 | 0.86 | 0.82 |
| B4GALNT4 | 0.85 | 0.82 |
| PLSCR3 | 0.85 | 0.81 |
| NAGK | 0.84 | 0.84 |
| ANKZF1 | 0.84 | 0.82 |
| RBM5 | 0.84 | 0.83 |
| QTRT1 | 0.84 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.64 | -0.71 |
| FBXO2 | -0.62 | -0.66 |
| CA4 | -0.62 | -0.75 |
| ALDOC | -0.62 | -0.70 |
| PTH1R | -0.61 | -0.68 |
| CSRP1 | -0.60 | -0.71 |
| TINAGL1 | -0.60 | -0.68 |
| C5orf53 | -0.60 | -0.70 |
| PTGDS | -0.60 | -0.69 |
| LDHD | -0.60 | -0.65 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003682 | chromatin binding | IEA | - | |
| GO:0003696 | satellite DNA binding | TAS | 10615123 | |
| GO:0005515 | protein binding | NAS | 12095912 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0030983 | mismatched DNA binding | IEA | - | |
| GO:0019237 | centromeric DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007130 | synaptonemal complex assembly | IEA | Synap (GO term level: 11) | - |
| GO:0007131 | reciprocal meiotic recombination | NAS | 12095912 | |
| GO:0007140 | male meiosis | IEA | - | |
| GO:0007144 | female meiosis I | IEA | - | |
| GO:0006298 | mismatch repair | IEA | - | |
| GO:0006298 | mismatch repair | NAS | 12095912 | |
| GO:0008104 | protein localization | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000793 | condensed chromosome | IEA | - | |
| GO:0000795 | synaptonemal complex | IEA | - | |
| GO:0005634 | nucleus | TAS | 10615123 | |
| GO:0005712 | chiasma | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MISMATCH REPAIR | 23 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOSIS | 116 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME MEIOTIC RECOMBINATION | 86 | 62 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| GNATENKO PLATELET SIGNATURE | 48 | 28 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| MATZUK MEIOTIC AND DNA REPAIR | 39 | 26 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOCYTE | 72 | 55 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| RAGHAVACHARI PLATELET SPECIFIC GENES | 70 | 46 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 2HR DN | 55 | 35 | All SZGR 2.0 genes in this pathway |