Gene Page: GP2
Summary ?
| GeneID | 2813 |
| Symbol | GP2 |
| Synonyms | ZAP75 |
| Description | glycoprotein 2 |
| Reference | MIM:602977|HGNC:HGNC:4441|Ensembl:ENSG00000169347|HPRD:04279|Vega:OTTHUMG00000131489 |
| Gene type | protein-coding |
| Map location | 16p12 |
| Pascal p-value | 0.304 |
| Fetal beta | -0.241 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
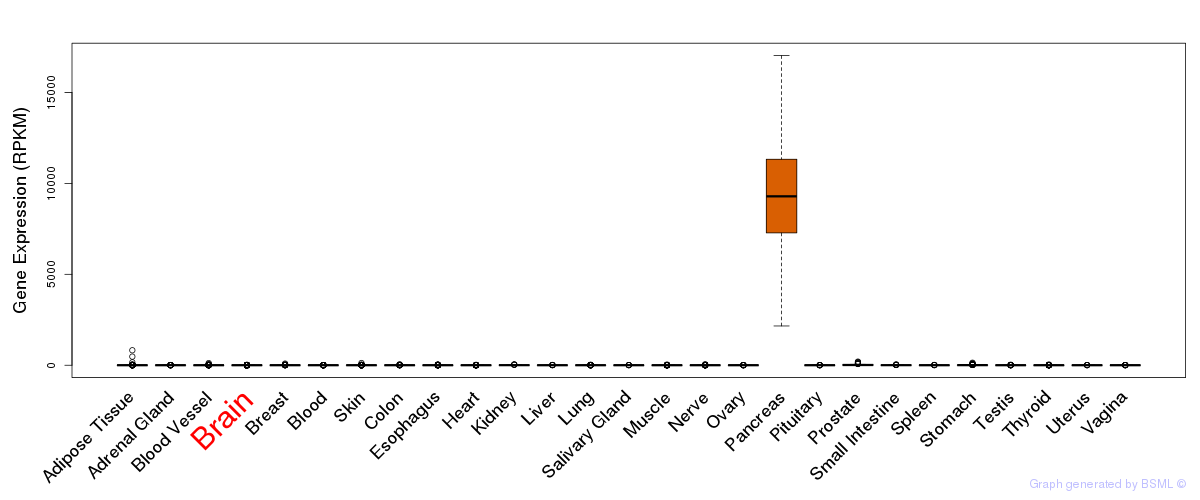
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DRD2 | 0.94 | 0.68 |
| GPR88 | 0.93 | 0.63 |
| DRD1 | 0.91 | 0.71 |
| DACT2 | 0.89 | 0.26 |
| AC020658.1 | 0.89 | 0.51 |
| PENK | 0.89 | 0.41 |
| SH3RF2 | 0.89 | 0.75 |
| ONECUT2 | 0.88 | 0.37 |
| HTR6 | 0.87 | 0.75 |
| RGS9 | 0.86 | 0.15 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CDADC1 | -0.23 | 0.16 |
| CCDC90A | -0.21 | -0.28 |
| RTN4RL2 | -0.20 | 0.28 |
| LRFN2 | -0.20 | 0.03 |
| EMX1 | -0.19 | -0.09 |
| IGSF21 | -0.19 | -0.04 |
| AC106818.2 | -0.19 | 0.08 |
| GTDC1 | -0.19 | 0.01 |
| GARNL3 | -0.19 | -0.09 |
| NXPH3 | -0.19 | -0.19 |
Section III. Gene Ontology annotation
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0031225 | anchored to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| VECCHI GASTRIC CANCER ADVANCED VS EARLY DN | 138 | 70 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| ROPERO HDAC2 TARGETS | 114 | 71 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP | 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS UP | 221 | 120 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |