Gene Page: HLA-DPB1
Summary ?
| GeneID | 3115 |
| Symbol | HLA-DPB1 |
| Synonyms | DPB1|HLA-DP|HLA-DP1B|HLA-DPB |
| Description | major histocompatibility complex, class II, DP beta 1 |
| Reference | MIM:142858|HGNC:HGNC:4940|HPRD:00832| |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 1.1E-5 |
| Sherlock p-value | 0.044 |
| Fetal beta | -1.06 |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11216730 | chr11 | 117918701 | HLA-DPB1 | 3115 | 0.13 | trans | ||
| rs9277376 | 6 | 33050196 | HLA-DPB1 | ENSG00000223865.6 | 9.011E-7 | 0.01 | 6493 | gtex_brain_putamen_basal |
| rs9277377 | 6 | 33050203 | HLA-DPB1 | ENSG00000223865.6 | 9.011E-7 | 0.01 | 6500 | gtex_brain_putamen_basal |
| rs9277392 | 6 | 33050829 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 7126 | gtex_brain_putamen_basal |
| rs9277393 | 6 | 33050877 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 7174 | gtex_brain_putamen_basal |
| rs9277394 | 6 | 33050970 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 7267 | gtex_brain_putamen_basal |
| rs9277395 | 6 | 33051051 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 7348 | gtex_brain_putamen_basal |
| rs9277396 | 6 | 33051139 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 7436 | gtex_brain_putamen_basal |
| rs9277401 | 6 | 33051394 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 7691 | gtex_brain_putamen_basal |
| rs9277402 | 6 | 33051460 | HLA-DPB1 | ENSG00000223865.6 | 3.218E-7 | 0.01 | 7757 | gtex_brain_putamen_basal |
| rs9277406 | 6 | 33051554 | HLA-DPB1 | ENSG00000223865.6 | 4.173E-7 | 0.01 | 7851 | gtex_brain_putamen_basal |
| rs9277407 | 6 | 33051558 | HLA-DPB1 | ENSG00000223865.6 | 4.173E-7 | 0.01 | 7855 | gtex_brain_putamen_basal |
| rs9277408 | 6 | 33051562 | HLA-DPB1 | ENSG00000223865.6 | 4.18E-7 | 0.01 | 7859 | gtex_brain_putamen_basal |
| rs9277409 | 6 | 33051624 | HLA-DPB1 | ENSG00000223865.6 | 4.116E-7 | 0.01 | 7921 | gtex_brain_putamen_basal |
| rs9277410 | 6 | 33051640 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 7937 | gtex_brain_putamen_basal |
| rs9277411 | 6 | 33051683 | HLA-DPB1 | ENSG00000223865.6 | 4.083E-7 | 0.01 | 7980 | gtex_brain_putamen_basal |
| rs9277412 | 6 | 33051689 | HLA-DPB1 | ENSG00000223865.6 | 4.083E-7 | 0.01 | 7986 | gtex_brain_putamen_basal |
| rs9277420 | 6 | 33051790 | HLA-DPB1 | ENSG00000223865.6 | 1.607E-7 | 0.01 | 8087 | gtex_brain_putamen_basal |
| rs9282411 | 6 | 33051827 | HLA-DPB1 | ENSG00000223865.6 | 1.609E-7 | 0.01 | 8124 | gtex_brain_putamen_basal |
| rs9277423 | 6 | 33051845 | HLA-DPB1 | ENSG00000223865.6 | 1.607E-7 | 0.01 | 8142 | gtex_brain_putamen_basal |
| rs9277424 | 6 | 33051865 | HLA-DPB1 | ENSG00000223865.6 | 1.607E-7 | 0.01 | 8162 | gtex_brain_putamen_basal |
| rs142975055 | 6 | 33051896 | HLA-DPB1 | ENSG00000223865.6 | 1.626E-7 | 0.01 | 8193 | gtex_brain_putamen_basal |
| rs9277426 | 6 | 33051910 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 8207 | gtex_brain_putamen_basal |
| rs9277427 | 6 | 33051969 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 8266 | gtex_brain_putamen_basal |
| rs9277428 | 6 | 33051976 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 8273 | gtex_brain_putamen_basal |
| rs9277431 | 6 | 33052028 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 8325 | gtex_brain_putamen_basal |
| rs9277432 | 6 | 33052072 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 8369 | gtex_brain_putamen_basal |
| rs9277433 | 6 | 33052128 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 8425 | gtex_brain_putamen_basal |
| rs10536191 | 6 | 33052167 | HLA-DPB1 | ENSG00000223865.6 | 3.168E-7 | 0.01 | 8464 | gtex_brain_putamen_basal |
| rs9277437 | 6 | 33052250 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 8547 | gtex_brain_putamen_basal |
| rs9277438 | 6 | 33052303 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 8600 | gtex_brain_putamen_basal |
| rs9277439 | 6 | 33052315 | HLA-DPB1 | ENSG00000223865.6 | 3.821E-7 | 0.01 | 8612 | gtex_brain_putamen_basal |
| rs9277440 | 6 | 33052327 | HLA-DPB1 | ENSG00000223865.6 | 3.715E-7 | 0.01 | 8624 | gtex_brain_putamen_basal |
| rs9277441 | 6 | 33052354 | HLA-DPB1 | ENSG00000223865.6 | 3.821E-7 | 0.01 | 8651 | gtex_brain_putamen_basal |
| rs9277443 | 6 | 33052414 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 8711 | gtex_brain_putamen_basal |
| rs9277444 | 6 | 33052439 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 8736 | gtex_brain_putamen_basal |
| rs9277445 | 6 | 33052492 | HLA-DPB1 | ENSG00000223865.6 | 2.168E-7 | 0.01 | 8789 | gtex_brain_putamen_basal |
| rs9277446 | 6 | 33052498 | HLA-DPB1 | ENSG00000223865.6 | 1.594E-7 | 0.01 | 8795 | gtex_brain_putamen_basal |
| rs1042187 | 6 | 33052768 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9065 | gtex_brain_putamen_basal |
| rs1042212 | 6 | 33052803 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9100 | gtex_brain_putamen_basal |
| rs1042331 | 6 | 33052950 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9247 | gtex_brain_putamen_basal |
| rs1042335 | 6 | 33052958 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9255 | gtex_brain_putamen_basal |
| rs1071597 | 6 | 33052986 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9283 | gtex_brain_putamen_basal |
| rs9277458 | 6 | 33053167 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9464 | gtex_brain_putamen_basal |
| rs9277459 | 6 | 33053248 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9545 | gtex_brain_putamen_basal |
| rs9277460 | 6 | 33053252 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9549 | gtex_brain_putamen_basal |
| rs9277461 | 6 | 33053270 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9567 | gtex_brain_putamen_basal |
| rs9277462 | 6 | 33053271 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9568 | gtex_brain_putamen_basal |
| rs9277463 | 6 | 33053307 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9604 | gtex_brain_putamen_basal |
| rs9277466 | 6 | 33053399 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9696 | gtex_brain_putamen_basal |
| rs9277467 | 6 | 33053444 | HLA-DPB1 | ENSG00000223865.6 | 9.748E-7 | 0.01 | 9741 | gtex_brain_putamen_basal |
| rs9277468 | 6 | 33053455 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9752 | gtex_brain_putamen_basal |
| rs9277469 | 6 | 33053468 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9765 | gtex_brain_putamen_basal |
| rs9277470 | 6 | 33053477 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9774 | gtex_brain_putamen_basal |
| rs9277471 | 6 | 33053682 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 9979 | gtex_brain_putamen_basal |
| rs9277481 | 6 | 33053820 | HLA-DPB1 | ENSG00000223865.6 | 3.394E-7 | 0.01 | 10117 | gtex_brain_putamen_basal |
| rs9277490 | 6 | 33053968 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 10265 | gtex_brain_putamen_basal |
| rs9277491 | 6 | 33053971 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 10268 | gtex_brain_putamen_basal |
| rs9277492 | 6 | 33053982 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 10279 | gtex_brain_putamen_basal |
| rs9277493 | 6 | 33054046 | HLA-DPB1 | ENSG00000223865.6 | 3.659E-7 | 0.01 | 10343 | gtex_brain_putamen_basal |
| rs9277494 | 6 | 33054057 | HLA-DPB1 | ENSG00000223865.6 | 3.428E-7 | 0.01 | 10354 | gtex_brain_putamen_basal |
| rs9277497 | 6 | 33054091 | HLA-DPB1 | ENSG00000223865.6 | 1.631E-7 | 0.01 | 10388 | gtex_brain_putamen_basal |
| rs9277511 | 6 | 33054215 | HLA-DPB1 | ENSG00000223865.6 | 3.912E-7 | 0.01 | 10512 | gtex_brain_putamen_basal |
| rs9277513 | 6 | 33054231 | HLA-DPB1 | ENSG00000223865.6 | 7.576E-7 | 0.01 | 10528 | gtex_brain_putamen_basal |
| rs9277514 | 6 | 33054235 | HLA-DPB1 | ENSG00000223865.6 | 7.648E-7 | 0.01 | 10532 | gtex_brain_putamen_basal |
| rs9277515 | 6 | 33054268 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 10565 | gtex_brain_putamen_basal |
| rs9277518 | 6 | 33054302 | HLA-DPB1 | ENSG00000223865.6 | 1.63E-7 | 0.01 | 10599 | gtex_brain_putamen_basal |
| rs1126719 | 6 | 33054325 | HLA-DPB1 | ENSG00000223865.6 | 2.189E-7 | 0.01 | 10622 | gtex_brain_putamen_basal |
| rs1126723 | 6 | 33054331 | HLA-DPB1 | ENSG00000223865.6 | 2.189E-7 | 0.01 | 10628 | gtex_brain_putamen_basal |
| rs1042544 | 6 | 33054457 | HLA-DPB1 | ENSG00000223865.6 | 1.631E-7 | 0.01 | 10754 | gtex_brain_putamen_basal |
| rs931 | 6 | 33054550 | HLA-DPB1 | ENSG00000223865.6 | 4.117E-7 | 0.01 | 10847 | gtex_brain_putamen_basal |
| rs928 | 6 | 33054552 | HLA-DPB1 | ENSG00000223865.6 | 4.117E-7 | 0.01 | 10849 | gtex_brain_putamen_basal |
| rs929 | 6 | 33054619 | HLA-DPB1 | ENSG00000223865.6 | 4.123E-7 | 0.01 | 10916 | gtex_brain_putamen_basal |
| rs9277532 | 6 | 33054711 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11008 | gtex_brain_putamen_basal |
| rs9277533 | 6 | 33054721 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11018 | gtex_brain_putamen_basal |
| rs9277534 | 6 | 33054807 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11104 | gtex_brain_putamen_basal |
| rs9277536 | 6 | 33054890 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11187 | gtex_brain_putamen_basal |
| rs9277537 | 6 | 33055009 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11306 | gtex_brain_putamen_basal |
| rs9277538 | 6 | 33055047 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11344 | gtex_brain_putamen_basal |
| rs9277540 | 6 | 33055123 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11420 | gtex_brain_putamen_basal |
| rs9277541 | 6 | 33055158 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11455 | gtex_brain_putamen_basal |
| rs9280312 | 6 | 33055196 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11493 | gtex_brain_putamen_basal |
| rs9277542 | 6 | 33055247 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11544 | gtex_brain_putamen_basal |
| rs9277544 | 6 | 33055308 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11605 | gtex_brain_putamen_basal |
| rs9277546 | 6 | 33055346 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11643 | gtex_brain_putamen_basal |
| rs9277547 | 6 | 33055367 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11664 | gtex_brain_putamen_basal |
| rs9277548 | 6 | 33055390 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11687 | gtex_brain_putamen_basal |
| rs9277549 | 6 | 33055419 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11716 | gtex_brain_putamen_basal |
| rs9277550 | 6 | 33055487 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11784 | gtex_brain_putamen_basal |
| rs9277551 | 6 | 33055494 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11791 | gtex_brain_putamen_basal |
| rs9277553 | 6 | 33055516 | HLA-DPB1 | ENSG00000223865.6 | 4.123E-7 | 0.01 | 11813 | gtex_brain_putamen_basal |
| rs9277554 | 6 | 33055538 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 11835 | gtex_brain_putamen_basal |
| rs3128963 | 6 | 33055780 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 12077 | gtex_brain_putamen_basal |
| rs3128964 | 6 | 33055818 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 12115 | gtex_brain_putamen_basal |
| rs5875436 | 6 | 33055843 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 12140 | gtex_brain_putamen_basal |
| rs9282413 | 6 | 33056396 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 12693 | gtex_brain_putamen_basal |
| rs3117228 | 6 | 33056435 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 12732 | gtex_brain_putamen_basal |
| rs9277566 | 6 | 33056916 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 13213 | gtex_brain_putamen_basal |
| rs9277568 | 6 | 33057055 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 13352 | gtex_brain_putamen_basal |
| rs3130188 | 6 | 33057176 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 13473 | gtex_brain_putamen_basal |
| rs3091282 | 6 | 33057198 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 13495 | gtex_brain_putamen_basal |
| rs534504368 | 6 | 33057511 | HLA-DPB1 | ENSG00000223865.6 | 5.184E-7 | 0.01 | 13808 | gtex_brain_putamen_basal |
| rs3117225 | 6 | 33057711 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 14008 | gtex_brain_putamen_basal |
| rs3097652 | 6 | 33057835 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 14132 | gtex_brain_putamen_basal |
| rs2295118 | 6 | 33060892 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 17189 | gtex_brain_putamen_basal |
| rs3117216 | 6 | 33063403 | HLA-DPB1 | ENSG00000223865.6 | 4.127E-7 | 0.01 | 19700 | gtex_brain_putamen_basal |
| rs3128930 | 6 | 33075666 | HLA-DPB1 | ENSG00000223865.6 | 5.179E-7 | 0.01 | 31963 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
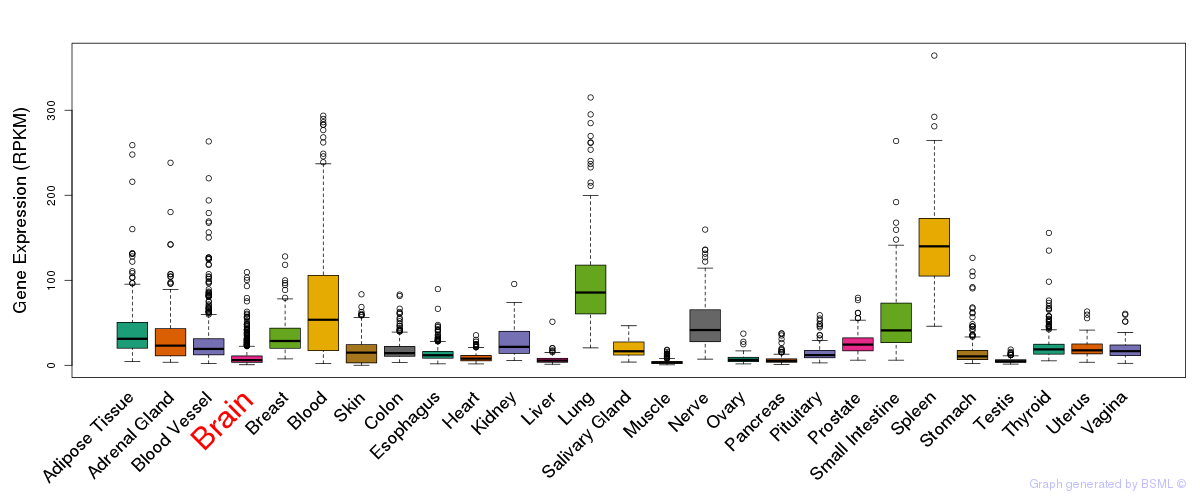
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GPER | 0.86 | 0.89 |
| SLC6A12 | 0.85 | 0.88 |
| MVP | 0.85 | 0.90 |
| PRELP | 0.84 | 0.89 |
| APOL3 | 0.84 | 0.86 |
| EPAS1 | 0.83 | 0.88 |
| GPR37L1 | 0.83 | 0.88 |
| TGM2 | 0.83 | 0.84 |
| PBXIP1 | 0.83 | 0.84 |
| BCAM | 0.82 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SUPV3L1 | -0.71 | -0.76 |
| MED19 | -0.71 | -0.76 |
| FRG1 | -0.71 | -0.78 |
| DUSP12 | -0.71 | -0.74 |
| ZNF821 | -0.70 | -0.70 |
| ZBED5 | -0.70 | -0.77 |
| STMN1 | -0.70 | -0.74 |
| GNL3 | -0.70 | -0.73 |
| PAK1IP1 | -0.69 | -0.73 |
| CYP2R1 | -0.69 | -0.77 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0032395 | MHC class II receptor activity | TAS | 8105536 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | IEA | - | |
| GO:0009405 | pathogenesis | TAS | 8105536 | |
| GO:0006955 | immune response | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 11048639 |11827988 |15489916 |17652306 | |
| GO:0042613 | MHC class II protein complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG INTESTINAL IMMUNE NETWORK FOR IGA PRODUCTION | 48 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE I DIABETES MELLITUS | 44 | 38 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ASTHMA | 30 | 25 | All SZGR 2.0 genes in this pathway |
| KEGG AUTOIMMUNE THYROID DISEASE | 53 | 49 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| KEGG ALLOGRAFT REJECTION | 38 | 34 | All SZGR 2.0 genes in this pathway |
| KEGG GRAFT VERSUS HOST DISEASE | 42 | 31 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME MHC CLASS II ANTIGEN PRESENTATION | 91 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME TCR SIGNALING | 54 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM TCR SIGNALING | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | 16 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | 14 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERATION OF SECOND MESSENGER MOLECULES | 27 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME PD1 SIGNALING | 18 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME COSTIMULATION BY THE CD28 FAMILY | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON GAMMA SIGNALING | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| GRAHAM CML QUIESCENT VS NORMAL QUIESCENT DN | 47 | 32 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 4 | 16 | 9 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| ROSS ACUTE MYELOID LEUKEMIA CBF | 82 | 57 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION L0 L1 DN | 21 | 14 | All SZGR 2.0 genes in this pathway |
| WIELAND UP BY HBV INFECTION | 101 | 66 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP | 114 | 84 | All SZGR 2.0 genes in this pathway |
| PETROVA ENDOTHELIUM LYMPHATIC VS BLOOD UP | 131 | 87 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP | 101 | 76 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES DN | 245 | 144 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| MCLACHLAN DENTAL CARIES UP | 253 | 147 | All SZGR 2.0 genes in this pathway |
| KIM LRRC3B TARGETS | 30 | 24 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS UP | 317 | 208 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL CARCINOMA VS ADENOMA DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION C | 69 | 49 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G5 DN | 27 | 17 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |