Gene Page: APOC3
Summary ?
| GeneID | 345 |
| Symbol | APOC3 |
| Synonyms | APOCIII|HALP2 |
| Description | apolipoprotein C-III |
| Reference | MIM:107720|HGNC:HGNC:610|Ensembl:ENSG00000110245|HPRD:00132|Vega:OTTHUMG00000046115 |
| Gene type | protein-coding |
| Map location | 11q23.3 |
| Pascal p-value | 0.618 |
| Fetal beta | 0.05 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
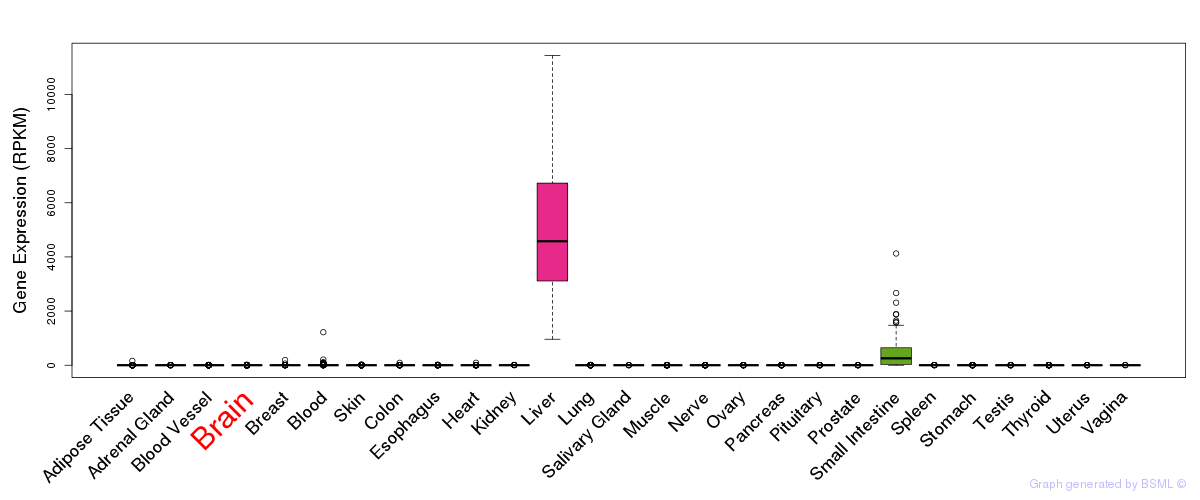
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CST3 | 0.91 | 0.96 |
| AC011427.1 | 0.86 | 0.93 |
| SLC7A10 | 0.85 | 0.91 |
| HSD17B14 | 0.84 | 0.91 |
| METRN | 0.82 | 0.91 |
| AIFM3 | 0.82 | 0.90 |
| TSC22D4 | 0.81 | 0.90 |
| S100A16 | 0.80 | 0.91 |
| GATSL3 | 0.79 | 0.76 |
| TLCD1 | 0.79 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C6orf168 | -0.75 | -0.83 |
| SPATS2 | -0.73 | -0.85 |
| EIF4ENIF1 | -0.73 | -0.81 |
| PREP | -0.72 | -0.81 |
| RASA1 | -0.72 | -0.79 |
| ARIH2 | -0.72 | -0.82 |
| TERF2 | -0.72 | -0.84 |
| ZNF329 | -0.72 | -0.83 |
| UBP1 | -0.72 | -0.79 |
| ZRANB1 | -0.72 | -0.82 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005543 | phospholipid binding | IDA | 4066713 |11060345 | |
| GO:0005319 | lipid transporter activity | IEA | - | |
| GO:0030234 | enzyme regulator activity | IDA | 11060345 | |
| GO:0015485 | cholesterol binding | IC | 11162594 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | IDA | 16443932 | |
| GO:0016042 | lipid catabolic process | IEA | - | |
| GO:0006869 | lipid transport | IEA | - | |
| GO:0033344 | cholesterol efflux | IDA | 11162594 |16443932 | |
| GO:0051005 | negative regulation of lipoprotein lipase activity | IDA | 11060345 | |
| GO:0042157 | lipoprotein metabolic process | IEA | - | |
| GO:0019433 | triacylglycerol catabolic process | IDA | 11060345 | |
| GO:0034375 | high-density lipoprotein particle remodeling | IMP | 17438339 | |
| GO:0032488 | Cdc42 protein signal transduction | IDA | 16443932 | |
| GO:0033700 | phospholipid efflux | IDA | 11162594 | |
| GO:0045717 | negative regulation of fatty acid biosynthetic process | IDA | 11060345 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | EXP | 4345202 | |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IDA | 11060345 | |
| GO:0034366 | spherical high-density lipoprotein particle | IDA | 17438339 | |
| GO:0034363 | intermediate-density lipoprotein particle | IDA | 17336988 | |
| GO:0034361 | very-low-density lipoprotein particle | IDA | 16935699 | |
| GO:0042627 | chylomicron | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PPAR SIGNALING PATHWAY | 69 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME HDL MEDIATED LIPID TRANSPORT | 15 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | 46 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPOPROTEIN METABOLISM | 28 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | 16 | 15 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| HOEGERKORP CD44 TARGETS TEMPORAL DN | 25 | 16 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE UP | 70 | 49 | All SZGR 2.0 genes in this pathway |
| WOO LIVER CANCER RECURRENCE DN | 80 | 54 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |