Gene Page: IL12A
Summary ?
| GeneID | 3592 |
| Symbol | IL12A |
| Synonyms | CLMF|IL-12A|NFSK|NKSF1|P35 |
| Description | interleukin 12A |
| Reference | MIM:161560|HGNC:HGNC:5969|Ensembl:ENSG00000168811|HPRD:01193|Vega:OTTHUMG00000158942 |
| Gene type | protein-coding |
| Map location | 3q25.33 |
| Pascal p-value | 0.698 |
| Fetal beta | -0.133 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
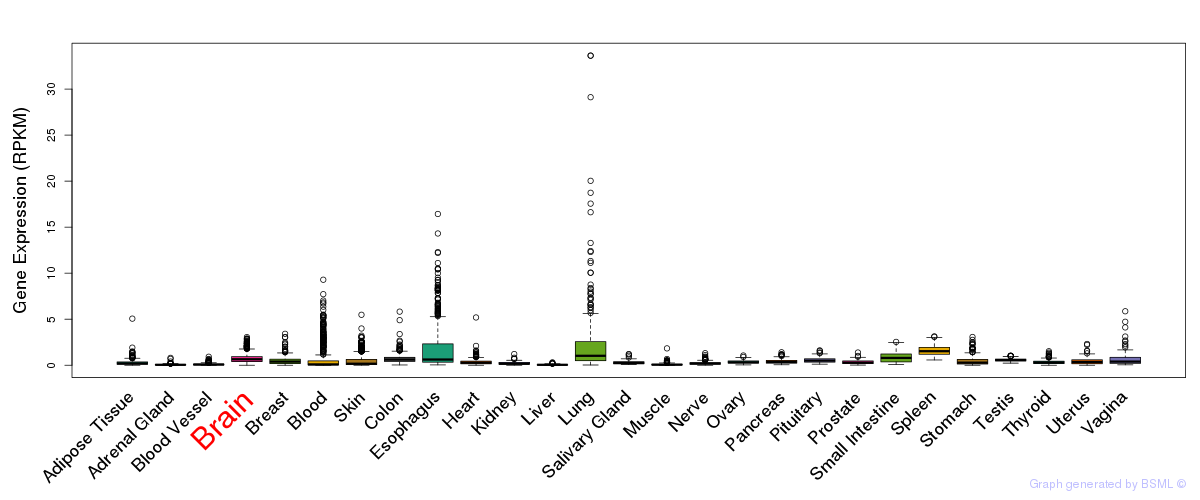
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TESC | 0.78 | 0.65 |
| AC005512.1 | 0.70 | 0.57 |
| TPD52L1 | 0.69 | 0.54 |
| HPCA | 0.69 | 0.57 |
| C20orf103 | 0.68 | 0.60 |
| PDE1B | 0.67 | 0.67 |
| LRPAP1 | 0.67 | 0.57 |
| TMEM90A | 0.66 | 0.30 |
| PPP1R1B | 0.66 | 0.18 |
| GSTO2 | 0.65 | 0.51 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SH2B2 | -0.29 | -0.50 |
| FADS2 | -0.29 | -0.40 |
| KIAA1949 | -0.29 | -0.41 |
| TUBB2B | -0.28 | -0.46 |
| TUBB | -0.28 | -0.26 |
| TUBB3 | -0.28 | -0.26 |
| MARCKSL1 | -0.28 | -0.34 |
| KIAA1211 | -0.28 | -0.29 |
| SH3BP2 | -0.28 | -0.45 |
| ZNF311 | -0.28 | -0.32 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005125 | cytokine activity | IDA | 1674604 | |
| GO:0005125 | cytokine activity | IEA | - | |
| GO:0005125 | cytokine activity | TAS | 1673147 | |
| GO:0005143 | interleukin-12 receptor binding | NAS | 1674604 | |
| GO:0005515 | protein binding | IPI | 10899108 | |
| GO:0042163 | interleukin-12 beta subunit binding | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IPI | 1674604 | |
| GO:0045513 | interleukin-27 binding | IPI | 9342359 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002860 | positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | IDA | 7903063 | |
| GO:0001916 | positive regulation of T cell mediated cytotoxicity | IDA | 1674604 |2204066 | |
| GO:0007050 | cell cycle arrest | IDA | 16942485 | |
| GO:0010224 | response to UV-B | IDA | 8992506 | |
| GO:0006955 | immune response | TAS | 9789052 | |
| GO:0032816 | positive regulation of natural killer cell activation | IDA | 1674604 | |
| GO:0016477 | cell migration | IDA | 7903063 | |
| GO:0042102 | positive regulation of T cell proliferation | IEA | - | |
| GO:0032729 | positive regulation of interferon-gamma production | IDA | 1674604 | |
| GO:0032496 | response to lipopolysaccharide | IDA | 7605994 |8557999 | |
| GO:0034393 | positive regulation of smooth muscle cell apoptosis | IDA | 16942485 | |
| GO:0042520 | positive regulation of tyrosine phosphorylation of Stat4 protein | IDA | 12372421 | |
| GO:0045785 | positive regulation of cell adhesion | IDA | 7903063 | |
| GO:0050830 | defense response to Gram-positive bacterium | IEP | 1357073 | |
| GO:0048662 | negative regulation of smooth muscle cell proliferation | IDA | 16942485 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0043514 | interleukin-12 complex | IDA | 2204066 | |
| GO:0043514 | interleukin-12 complex | IEA | - | |
| GO:0043514 | interleukin-12 complex | NAS | 1674604 |14718574 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE I DIABETES MELLITUS | 44 | 38 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ALLOGRAFT REJECTION | 38 | 34 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CYTOKINE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INFLAM PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DC PATHWAY | 22 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL12 PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NO2IL12 PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKT PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TH1TH2 PATHWAY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| PID IL27 PATHWAY | 26 | 20 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP | 233 | 161 | All SZGR 2.0 genes in this pathway |
| DEBOSSCHER NFKB TARGETS REPRESSED BY GLUCOCORTICOIDS | 24 | 18 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE UP | 86 | 57 | All SZGR 2.0 genes in this pathway |
| WANG TARGETS OF MLL CBP FUSION UP | 44 | 26 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING NOT VIA ATM UP | 62 | 29 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS UP | 165 | 118 | All SZGR 2.0 genes in this pathway |
| BUDHU LIVER CANCER METASTASIS DN | 7 | 7 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS HCP WITH H3 UNMETHYLATED | 80 | 50 | All SZGR 2.0 genes in this pathway |
| SEIKE LUNG CANCER POOR SURVIVAL | 11 | 8 | All SZGR 2.0 genes in this pathway |
| GHANDHI DIRECT IRRADIATION UP | 110 | 68 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-21 | 256 | 263 | 1A,m8 | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.