Gene Page: IL12B
Summary ?
| GeneID | 3593 |
| Symbol | IL12B |
| Synonyms | CLMF|CLMF2|IL-12B|IMD28|IMD29|NKSF|NKSF2 |
| Description | interleukin 12B |
| Reference | MIM:161561|HGNC:HGNC:5970|Ensembl:ENSG00000113302|HPRD:01194|Vega:OTTHUMG00000130307 |
| Gene type | protein-coding |
| Map location | 5q33.3 |
| Pascal p-value | 0.013 |
| Fetal beta | 0.182 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4864466 | chr4 | 54403975 | IL12B | 3593 | 0.11 | trans |
Section II. Transcriptome annotation
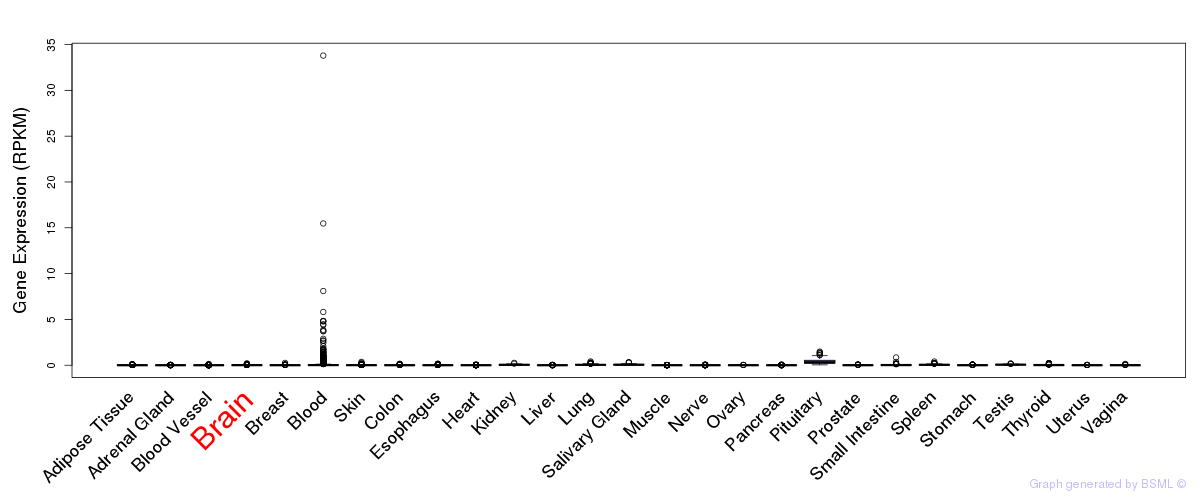
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | TAS | 9789052 | |
| GO:0005125 | cytokine activity | IDA | 1674604 | |
| GO:0004896 | cytokine receptor activity | IEA | - | |
| GO:0005143 | interleukin-12 receptor binding | TAS | 1674604 | |
| GO:0005515 | protein binding | IPI | 10899108 | |
| GO:0042164 | interleukin-12 alpha subunit binding | IEA | - | |
| GO:0042803 | protein homodimerization activity | IEA | - | |
| GO:0046982 | protein heterodimerization activity | IPI | 1674604 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002860 | positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | IDA | 7903063 | |
| GO:0007166 | cell surface receptor linked signal transduction | IEA | - | |
| GO:0007050 | cell cycle arrest | IDA | 16942485 | |
| GO:0010224 | response to UV-B | IDA | 8992506 | |
| GO:0042093 | T-helper cell differentiation | IDA | 1673147 | |
| GO:0032816 | positive regulation of natural killer cell activation | IDA | 1674604 | |
| GO:0016477 | cell migration | IDA | 7903063 | |
| GO:0042104 | positive regulation of activated T cell proliferation | IDA | 1674604 | |
| GO:0032729 | positive regulation of interferon-gamma production | IDA | 1674604 | |
| GO:0042088 | T-helper 1 type immune response | TAS | 1673147 | |
| GO:0034393 | positive regulation of smooth muscle cell apoptosis | IDA | 16942485 | |
| GO:0042035 | regulation of cytokine biosynthetic process | TAS | 1673147 | |
| GO:0045785 | positive regulation of cell adhesion | IDA | 7903063 | |
| GO:0045078 | positive regulation of interferon-gamma biosynthetic process | TAS | 1673147 | |
| GO:0048662 | negative regulation of smooth muscle cell proliferation | IDA | 16942485 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0043514 | interleukin-12 complex | IDA | 1674604 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE I DIABETES MELLITUS | 44 | 38 | All SZGR 2.0 genes in this pathway |
| KEGG LEISHMANIA INFECTION | 72 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG ALLOGRAFT REJECTION | 38 | 34 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CYTOKINE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INFLAM PATHWAY | 29 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DC PATHWAY | 22 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL12 PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NO2IL12 PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NKT PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TH1TH2 PATHWAY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| PID IL27 PATHWAY | 26 | 20 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID IL23 PATHWAY | 37 | 30 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| LIU TARGETS OF VMYB VS CMYB DN | 43 | 30 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| GAURNIER PSMD4 TARGETS | 73 | 55 | All SZGR 2.0 genes in this pathway |
| VILIMAS NOTCH1 TARGETS UP | 52 | 41 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION A | 67 | 52 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| BUDHU LIVER CANCER METASTASIS DN | 7 | 7 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR EVASION AND TOLEROGENICITY UP | 30 | 19 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-153 | 362 | 368 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-448 | 362 | 368 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.