Gene Page: NQO2
Summary ?
| GeneID | 4835 |
| Symbol | NQO2 |
| Synonyms | DHQV|DIA6|NMOR2|QR2 |
| Description | NAD(P)H dehydrogenase, quinone 2 |
| Reference | MIM:160998|HGNC:HGNC:7856|Ensembl:ENSG00000124588|HPRD:01190|Vega:OTTHUMG00000014130 |
| Gene type | protein-coding |
| Map location | 6p25.2 |
| Pascal p-value | 0.144 |
| Sherlock p-value | 0.5 |
| Fetal beta | -1.483 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26943189 | 6 | 3002278 | NQO2 | 1.24E-5 | -0.546 | 0.014 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9405188 | 6 | 2997544 | NQO2 | ENSG00000124588.15 | 5.08434E-9 | 0 | 3379 | gtex_brain_putamen_basal |
| rs6596927 | 6 | 2998319 | NQO2 | ENSG00000124588.15 | 1.45769E-6 | 0 | 4154 | gtex_brain_putamen_basal |
| rs2071002 | 6 | 3000303 | NQO2 | ENSG00000124588.15 | 2.07243E-7 | 0 | 6138 | gtex_brain_putamen_basal |
| rs9378754 | 6 | 3000832 | NQO2 | ENSG00000124588.15 | 1.67035E-6 | 0 | 6667 | gtex_brain_putamen_basal |
| rs9378755 | 6 | 3000978 | NQO2 | ENSG00000124588.15 | 1.56508E-9 | 0 | 6813 | gtex_brain_putamen_basal |
| rs9392449 | 6 | 3001431 | NQO2 | ENSG00000124588.15 | 1.4107E-7 | 0 | 7266 | gtex_brain_putamen_basal |
| rs138616686 | 6 | 3003970 | NQO2 | ENSG00000124588.15 | 8.53875E-8 | 0 | 9805 | gtex_brain_putamen_basal |
| rs4149360 | 6 | 3006807 | NQO2 | ENSG00000124588.15 | 1.04308E-7 | 0 | 12642 | gtex_brain_putamen_basal |
| rs4149363 | 6 | 3007478 | NQO2 | ENSG00000124588.15 | 2.47705E-7 | 0 | 13313 | gtex_brain_putamen_basal |
| rs62392497 | 6 | 3017861 | NQO2 | ENSG00000124588.15 | 1.60818E-6 | 0 | 23696 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
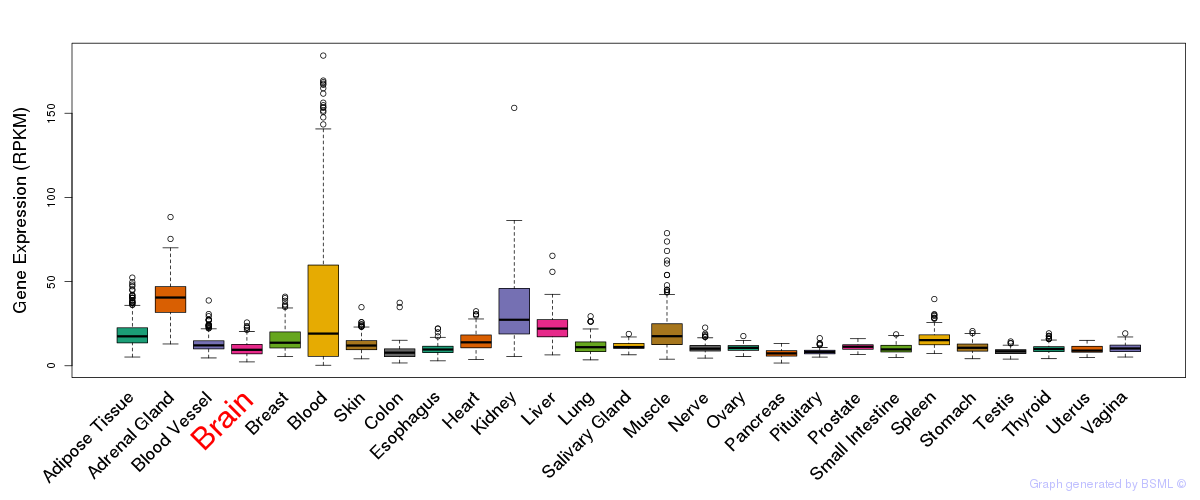
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001512 | dihydronicotinamide riboside quinone reductase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0008753 | NADPH dehydrogenase (quinone) activity | TAS | 1691923 | |
| GO:0009055 | electron carrier activity | IEA | - | |
| GO:0009055 | electron carrier activity | TAS | 1691923 | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0050662 | coenzyme binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0055114 | oxidation reduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| HALMOS CEBPA TARGETS UP | 52 | 34 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART VENTRICLE DN | 41 | 28 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 2D UP | 69 | 46 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER STEM CELL DN | 76 | 51 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA DN | 267 | 160 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| LI DCP2 BOUND MRNA | 89 | 57 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS UP | 61 | 41 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP | 279 | 155 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |