Gene Page: NPY1R
Summary ?
| GeneID | 4886 |
| Symbol | NPY1R |
| Synonyms | NPY1-R|NPYR |
| Description | neuropeptide Y receptor Y1 |
| Reference | MIM:162641|HGNC:HGNC:7956|Ensembl:ENSG00000164128|HPRD:01215|Vega:OTTHUMG00000161371 |
| Gene type | protein-coding |
| Map location | 4q32.2 |
| Pascal p-value | 0.362 |
| Fetal beta | -0.49 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24157598 | 4 | 164254508 | NPY1R | 3.026E-4 | 0.572 | 0.04 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| snp_a-2169623 | 0 | NPY1R | 4886 | 0.09 | trans | |||
| rs16894557 | chr6 | 28999825 | NPY1R | 4886 | 0.04 | trans |
Section II. Transcriptome annotation
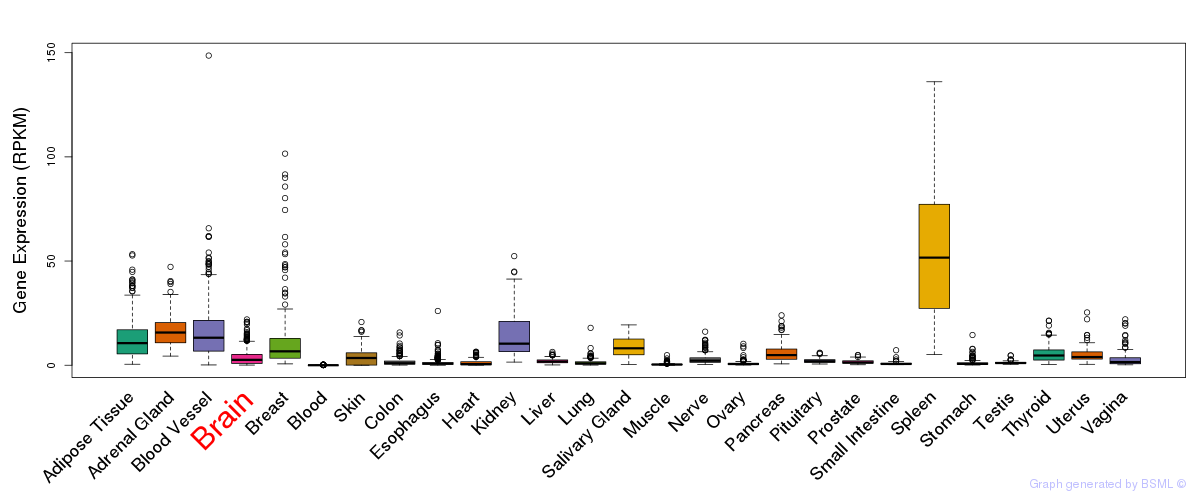
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM155B | 0.87 | 0.79 |
| CBFA2T3 | 0.85 | 0.77 |
| TNRC18B | 0.85 | 0.89 |
| KIRREL | 0.84 | 0.84 |
| AL139010.2 | 0.84 | 0.83 |
| BAHCC1 | 0.83 | 0.84 |
| ZNF512B | 0.82 | 0.83 |
| CACNA1H | 0.82 | 0.81 |
| ZNF710 | 0.82 | 0.82 |
| ZBED1 | 0.82 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.57 | -0.72 |
| ACOT13 | -0.53 | -0.73 |
| S100B | -0.53 | -0.71 |
| ACYP2 | -0.53 | -0.68 |
| CA4 | -0.52 | -0.66 |
| AF347015.31 | -0.52 | -0.72 |
| COPZ2 | -0.52 | -0.67 |
| HLA-F | -0.52 | -0.60 |
| AF347015.27 | -0.52 | -0.71 |
| B2M | -0.51 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001601 | peptide YY receptor activity | IEA | - | |
| GO:0001602 | pancreatic polypeptide receptor activity | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 15231747 | |
| GO:0004983 | neuropeptide Y receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007218 | neuropeptide signaling pathway | IEA | Neurotransmitter (GO term level: 8) | - |
| GO:0007187 | G-protein signaling, coupled to cyclic nucleotide second messenger | TAS | 1317848 | |
| GO:0008217 | regulation of blood pressure | IEA | - | |
| GO:0007193 | G-protein signaling, adenylate cyclase inhibiting pathway | TAS | 1321422 | |
| GO:0006006 | glucose metabolic process | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007631 | feeding behavior | IEA | - | |
| GO:0007626 | locomotory behavior | IEA | - | |
| GO:0019233 | sensory perception of pain | IEA | - | |
| GO:0040014 | regulation of multicellular organism growth | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 1317848 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| WILCOX RESPONSE TO PROGESTERONE UP | 152 | 90 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C DN | 19 | 11 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| WATTEL AUTONOMOUS THYROID ADENOMA DN | 55 | 29 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| STAEGE EWING FAMILY TUMOR | 33 | 22 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX DN | 80 | 49 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| SASSON FSH RESPONSE | 9 | 6 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER HCP WITH H3K27ME3 | 97 | 72 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN | 258 | 160 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| MASRI RESISTANCE TO TAMOXIFEN AND AROMATASE INHIBITORS UP | 20 | 10 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B UP | 172 | 109 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER ERBB2 DN | 5 | 5 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL UP | 260 | 174 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K4ME3 AND H3K27ME3 | 38 | 34 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 DN | 80 | 51 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 3 | 20 | 9 | All SZGR 2.0 genes in this pathway |
| PEDERSEN TARGETS OF 611CTF ISOFORM OF ERBB2 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-218 | 45 | 52 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-381 | 1090 | 1097 | 1A,m8 | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.