Gene Page: NTF3
Summary ?
| GeneID | 4908 |
| Symbol | NTF3 |
| Synonyms | HDNF|NGF-2|NGF2|NT-3|NT3 |
| Description | neurotrophin 3 |
| Reference | MIM:162660|HGNC:HGNC:8023|Ensembl:ENSG00000185652|HPRD:01219|Vega:OTTHUMG00000168649 |
| Gene type | protein-coding |
| Map location | 12p13 |
| Pascal p-value | 0.008 |
| Fetal beta | 2.551 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 9 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
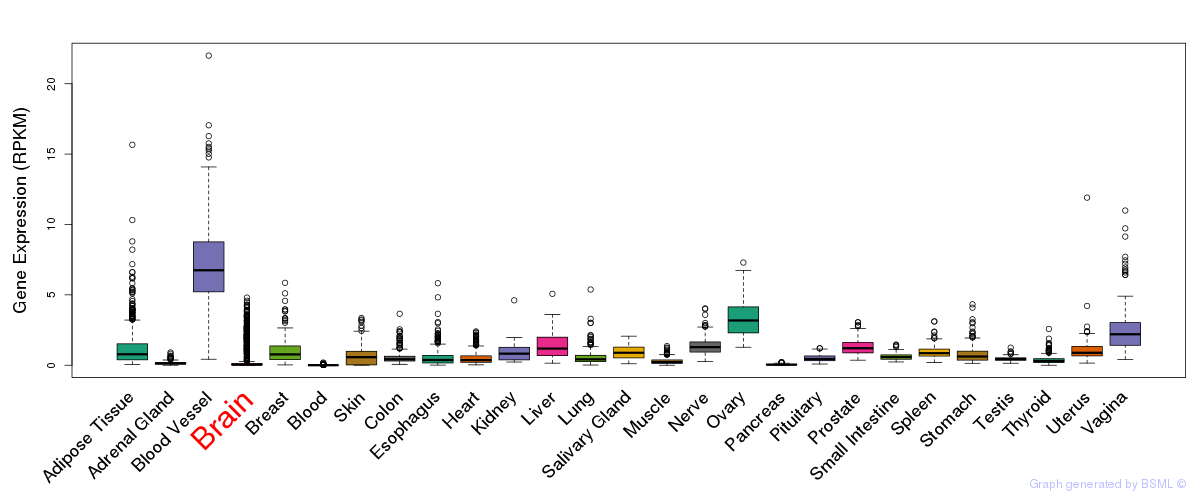
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RSAD2 | 0.85 | 0.78 |
| OAS3 | 0.83 | 0.79 |
| GBP1 | 0.80 | 0.77 |
| MX2 | 0.80 | 0.71 |
| PARP12 | 0.75 | 0.73 |
| HLA-B | 0.74 | 0.66 |
| SLC15A3 | 0.74 | 0.66 |
| ACSL5 | 0.73 | 0.75 |
| HLA-A | 0.73 | 0.57 |
| GBP3 | 0.72 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C17orf48 | -0.45 | -0.55 |
| RBMX2 | -0.45 | -0.56 |
| ZBED5 | -0.44 | -0.52 |
| RBM3 | -0.44 | -0.51 |
| MED19 | -0.44 | -0.54 |
| ZNF821 | -0.44 | -0.49 |
| WDR70 | -0.44 | -0.47 |
| BLVRA | -0.43 | -0.50 |
| SAAL1 | -0.43 | -0.51 |
| U2AF1 | -0.43 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005165 | neurotrophin receptor binding | IEA | - | |
| GO:0008083 | growth factor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | IEA | Brain (GO term level: 7) | - |
| GO:0007411 | axon guidance | IEA | axon (GO term level: 13) | - |
| GO:0007403 | glial cell fate determination | IEA | Glial (GO term level: 10) | - |
| GO:0048699 | generation of neurons | IEA | neuron, neurogenesis (GO term level: 7) | - |
| GO:0048666 | neuron development | IEA | neuron (GO term level: 9) | - |
| GO:0007274 | neuromuscular synaptic transmission | IEA | neuron, Synap (GO term level: 7) | - |
| GO:0048484 | enteric nervous system development | IEA | neuron (GO term level: 7) | - |
| GO:0042490 | mechanoreceptor differentiation | IEA | neuron (GO term level: 9) | - |
| GO:0043523 | regulation of neuron apoptosis | IEA | neuron (GO term level: 8) | - |
| GO:0007267 | cell-cell signaling | TAS | 2236018 | |
| GO:0007165 | signal transduction | TAS | 2236018 | |
| GO:0008544 | epidermis development | IEA | - | |
| GO:0007422 | peripheral nervous system development | IEA | - | |
| GO:0021675 | nerve development | IEA | - | |
| GO:0051145 | smooth muscle cell differentiation | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0016023 | cytoplasmic membrane-bounded vesicle | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER DN | 38 | 13 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL CURED VS FATAL UP | 39 | 22 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS UP | 214 | 133 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D2 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS UP | 135 | 96 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-200bc/429 | 249 | 255 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC | ||||
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-21 | 133 | 139 | m8 | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-221/222 | 52 | 59 | 1A,m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-369-3p | 85 | 91 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 85 | 92 | 1A,m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-410 | 80 | 86 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-448 | 46 | 52 | m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-539 | 142 | 148 | 1A | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.