Gene Page: NTRK1
Summary ?
| GeneID | 4914 |
| Symbol | NTRK1 |
| Synonyms | MTC|TRK|TRK1|TRKA|Trk-A|p140-TrkA |
| Description | neurotrophic tyrosine kinase, receptor, type 1 |
| Reference | MIM:191315|HGNC:HGNC:8031|Ensembl:ENSG00000198400|HPRD:01869|Vega:OTTHUMG00000041304 |
| Gene type | protein-coding |
| Map location | 1q21-q22 |
| Fetal beta | 0.09 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
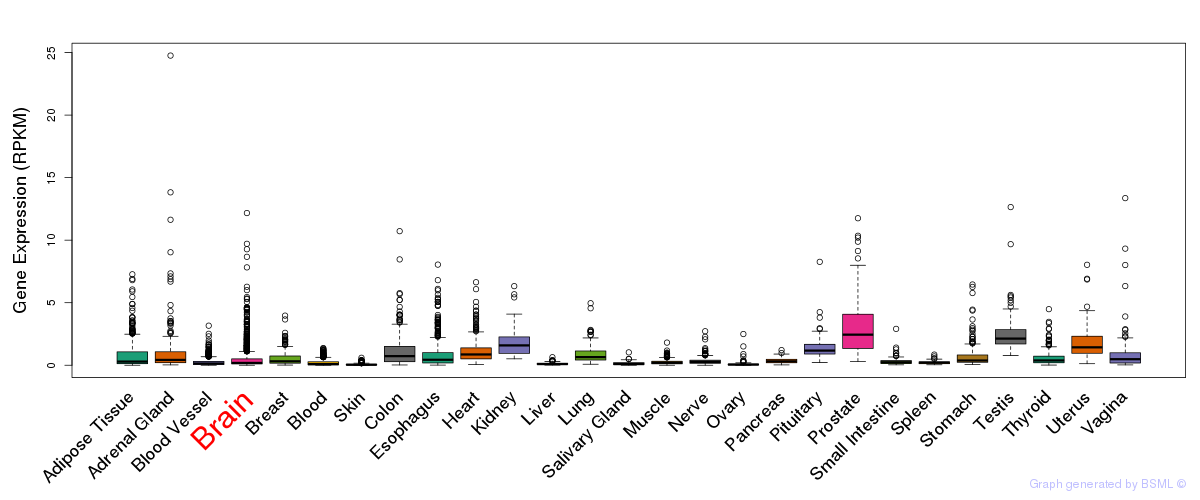
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | NAS | neurite (GO term level: 8) | 1695324 |
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | TAS | neurite (GO term level: 8) | 1695324 |8957089 |
| GO:0043121 | neurotrophin binding | TAS | neuron (GO term level: 5) | 9290260 |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0007190 | activation of adenylate cyclase activity | EXP | 16805430 | |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | EXP | 16939974 | |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | TAS | 9290260 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0006468 | protein amino acid phosphorylation | NAS | - | |
| GO:0007265 | Ras protein signal transduction | EXP | 11520933 | |
| GO:0008150 | biological_process | ND | 1695324 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | 1695324 | |
| GO:0005768 | endosome | EXP | 7806500 |9182757 |10629055 |16284401 | |
| GO:0016021 | integral to membrane | NAS | - | |
| GO:0005886 | plasma membrane | EXP | 11157096 |16284401 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9290260 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 10679771 |10708759 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 9867838 |
| FRS2 | FRS2A | FRS2alpha | SNT | SNT-1 | SNT1 | fibroblast growth factor receptor substrate 2 | TrkA interacts with FRS2-alpha. This interaction was modelled on a demonstrated interaction between human TrkA and mouse FRS2-alpha. | BIND | 10629055 |
| FRS2 | FRS2A | FRS2alpha | SNT | SNT-1 | SNT1 | fibroblast growth factor receptor substrate 2 | - | HPRD,BioGRID | 10092678 |
| FRS3 | FRS2B | FRS2beta | MGC17167 | SNT-2 | SNT2 | fibroblast growth factor receptor substrate 3 | TrkA interacts with FRS2-beta. | BIND | 10629055 |
| GIPC1 | C19orf3 | GIPC | GLUT1CBP | Hs.6454 | IIP-1 | MGC15889 | MGC3774 | NIP | RGS19IP1 | SEMCAP | SYNECTIIN | SYNECTIN | TIP-2 | GIPC PDZ domain containing family, member 1 | GIPC interacts with TRKA. This interaction was modelled on a demonstrated interaction between mouse GIPC and rat TrkA. | BIND | 11251075 |
| GIPC1 | C19orf3 | GIPC | GLUT1CBP | Hs.6454 | IIP-1 | MGC15889 | MGC3774 | NIP | RGS19IP1 | SEMCAP | SYNECTIIN | SYNECTIN | TIP-2 | GIPC PDZ domain containing family, member 1 | - | HPRD,BioGRID | 11251075 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 10748052 |11733534 |
| KIDINS220 | ARMS | MGC163482 | kinase D-interacting substrate, 220kDa | - | HPRD | 11150334 |15167895 |
| MATK | CHK | CTK | DKFZp434N1212 | HHYLTK | HYL | HYLTK | Lsk | MGC1708 | MGC2101 | megakaryocyte-associated tyrosine kinase | - | HPRD,BioGRID | 10329710 |
| NGF | Beta-NGF | HSAN5 | MGC161426 | MGC161428 | NGFB | nerve growth factor (beta polypeptide) | Interaction between TrkA (PDB ID: 1WWW_Y) and NGF (PDB ID: 1WWW_W). | BIND | 10490030 |
| NGF | Beta-NGF | HSAN5 | MGC161426 | MGC161428 | NGFB | nerve growth factor (beta polypeptide) | NGF interacts with TrkA. This interaction was modeled on a demonstrated interaction between NGF from human and TrkA from an unspecified species. | BIND | 14985763 |
| NGF | Beta-NGF | HSAN5 | MGC161426 | MGC161428 | NGFB | nerve growth factor (beta polypeptide) | Interaction between TrkA (PDB ID: 1WWW_Y) and NGF (PDB ID: 1WWW_V). | BIND | 10490030 |
| NGF | Beta-NGF | HSAN5 | MGC161426 | MGC161428 | NGFB | nerve growth factor (beta polypeptide) | - | HPRD,BioGRID | 14985763 |
| NGF | Beta-NGF | HSAN5 | MGC161426 | MGC161428 | NGFB | nerve growth factor (beta polypeptide) | Interaction between TrkA (PDB ID: 1WWW_X) and NGF (PDB ID: 1WWW_V). | BIND | 10490030 |
| NGF | Beta-NGF | HSAN5 | MGC161426 | MGC161428 | NGFB | nerve growth factor (beta polypeptide) | Interaction between TrkA (PDB ID: 1WWW_X) and NGF (PDB ID: 1WWW_W). | BIND | 10490030 |
| NTF3 | HDNF | MGC129711 | NGF-2 | NGF2 | NT3 | neurotrophin 3 | Reconstituted Complex | BioGRID | 8621424 |
| NTRK1 | DKFZp781I14186 | MTC | TRK | TRK1 | TRKA | p140-TrkA | neurotrophic tyrosine kinase, receptor, type 1 | - | HPRD,BioGRID | 10388563 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 1715690 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD,BioGRID | 12237455 |
| RAP1A | KREV-1 | KREV1 | RAP1 | SMGP21 | RAP1A, member of RAS oncogene family | Affinity Capture-Western | BioGRID | 11466412 |
| RGS19 | GAIP | RGSGAIP | regulator of G-protein signaling 19 | Affinity Capture-Western | BioGRID | 11251075 |
| RICS | GC-GAP | GRIT | KIAA0712 | MGC1892 | p200RhoGAP | p250GAP | Rho GTPase-activating protein | - | HPRD,BioGRID | 12446789 |
| RUSC1 | DKFZp761A1822 | NESCA | RUN and SH3 domain containing 1 | - | HPRD | 15024033 |
| SH2B1 | DKFZp547G1110 | FLJ30542 | KIAA1299 | SH2-B | SH2B | SH2B adaptor protein 1 | - | HPRD | 10708759 |
| SH2B1 | DKFZp547G1110 | FLJ30542 | KIAA1299 | SH2-B | SH2B | SH2B adaptor protein 1 | - | HPRD,BioGRID | 9856458 |
| SH2B2 | APS | SH2B adaptor protein 2 | Affinity Capture-Western Two-hybrid | BioGRID | 9856458 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Shc interacts with Trk. This interaction was modelled on a demonstrated interaction between SHC from rat or mouse and Trk from human. | BIND | 8155326 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Shc interacts with NGFR. | BIND | 8577769 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | ShcA interacts with TrkA. | BIND | 7542991 |8610109 |8943228 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 8183561 |9856458 |10092678 |10708759 |12446789 |
| SHC3 | N-Shc | NSHC | RAI | SHCC | SHC (Src homology 2 domain containing) transforming protein 3 | - | HPRD,BioGRID | 9507002 |
| SQSTM1 | A170 | OSIL | PDB3 | ZIP3 | p60 | p62 | p62B | sequestosome 1 | Affinity Capture-Western | BioGRID | 11244088 |12471037 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG THYROID CANCER | 29 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ERK5 PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TRKA PATHWAY | 12 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ACTINY PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| ST DIFFERENTIATION PATHWAY IN PC12 CELLS | 45 | 35 | All SZGR 2.0 genes in this pathway |
| SA TRKA RECEPTOR | 17 | 15 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID SHP2 PATHWAY | 58 | 46 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| PID PI3K PLC TRK PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME ARMS MEDIATED ACTIVATION | 17 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME PROLONGED ERK ACTIVATION EVENTS | 19 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO ERKS | 36 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO P38 VIA RIT AND RIN | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | 13 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K AKT ACTIVATION | 38 | 29 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| TANAKA METHYLATED IN ESOPHAGEAL CARCINOMA | 103 | 58 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 17 | 25 | 12 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 24 | 14 | 7 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| PETRETTO BLOOD PRESSURE UP | 12 | 8 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED SIGNIFICANTLY | 26 | 22 | All SZGR 2.0 genes in this pathway |
| RADAEVA RESPONSE TO IFNA1 UP | 52 | 40 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| ASGHARZADEH NEUROBLASTOMA POOR SURVIVAL DN | 46 | 30 | All SZGR 2.0 genes in this pathway |