Gene Page: ATP5B
Summary ?
| GeneID | 506 |
| Symbol | ATP5B |
| Synonyms | ATPMB|ATPSB|HEL-S-271 |
| Description | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| Reference | MIM:102910|HGNC:HGNC:830|Ensembl:ENSG00000110955|HPRD:00044| |
| Gene type | protein-coding |
| Map location | 12q13.13 |
| Pascal p-value | 0.33 |
| Sherlock p-value | 0.041 |
| Fetal beta | -0.437 |
| DMG | 1 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_mitochondria G2Cdb.human_Synaptosome G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0296 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17616707 | 12 | 57039822 | ATP5B | 6.11E-10 | -0.011 | 9.15E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
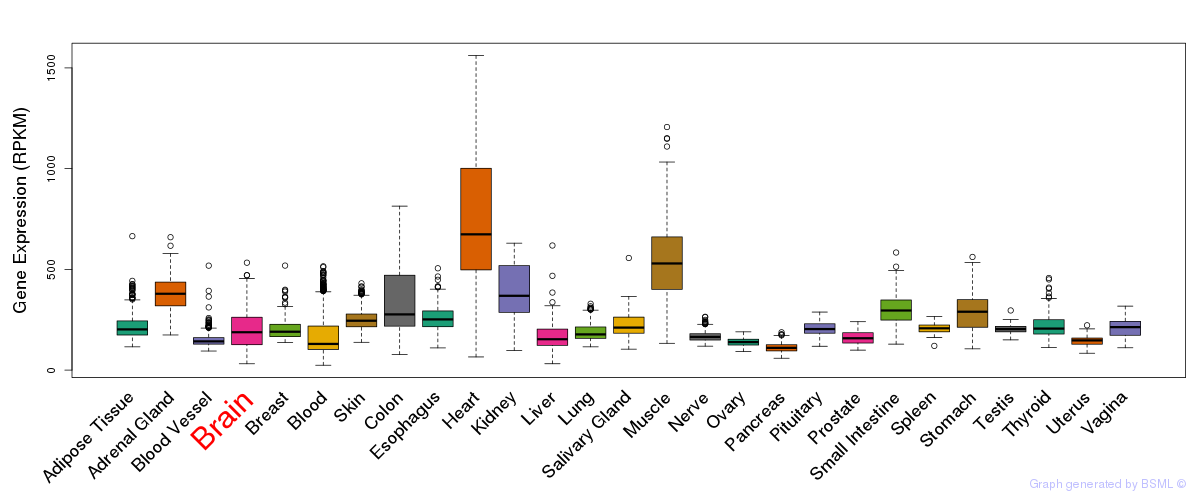
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SUCLG1 | 0.94 | 0.91 |
| PSMA6 | 0.92 | 0.91 |
| MRPL1 | 0.92 | 0.93 |
| ZNHIT3 | 0.92 | 0.90 |
| MRPL15 | 0.91 | 0.90 |
| CHMP5 | 0.90 | 0.88 |
| NFU1 | 0.90 | 0.91 |
| VPS29 | 0.89 | 0.91 |
| MRPL47 | 0.89 | 0.89 |
| C11orf73 | 0.89 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM38A | -0.57 | -0.62 |
| PPP1R13L | -0.56 | -0.55 |
| AF347015.26 | -0.55 | -0.53 |
| TRIM56 | -0.55 | -0.59 |
| AF347015.18 | -0.55 | -0.50 |
| DOCK6 | -0.54 | -0.56 |
| MYH9 | -0.54 | -0.53 |
| AF347015.2 | -0.54 | -0.51 |
| RRBP1 | -0.53 | -0.56 |
| TENC1 | -0.52 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 11410595 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0005215 | transporter activity | NAS | 2870059 | |
| GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism | IEA | - | |
| GO:0016887 | ATPase activity | IDA | 12110673 | |
| GO:0042288 | MHC class I protein binding | IDA | 17643490 | |
| GO:0043499 | eukaryotic cell surface binding | IDA | 10077593 | |
| GO:0046933 | hydrogen ion transporting ATP synthase activity, rotational mechanism | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0046961 | hydrogen ion transporting ATPase activity, rotational mechanism | IMP | 17510399 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001525 | angiogenesis | IMP | 17510399 | |
| GO:0006091 | generation of precursor metabolites and energy | NAS | 2870059 | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0015992 | proton transport | IMP | 17510399 | |
| GO:0015986 | ATP synthesis coupled proton transport | IEA | - | |
| GO:0051453 | regulation of intracellular pH | IMP | 17510399 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005739 | mitochondrion | IDA | 2870059 | |
| GO:0005753 | mitochondrial proton-transporting ATP synthase complex | IDA | 12110673 | |
| GO:0005753 | mitochondrial proton-transporting ATP synthase complex | NAS | 2900241 | |
| GO:0005754 | mitochondrial proton-transporting ATP synthase, catalytic core | NAS | 2687158 |2870059 | |
| GO:0005759 | mitochondrial matrix | EXP | 4517936 | |
| GO:0005759 | mitochondrial matrix | NAS | 2687158 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0009986 | cell surface | IDA | 17510399 | |
| GO:0005886 | plasma membrane | IDA | 10077593 | |
| GO:0016469 | proton-transporting two-sector ATPase complex | IEA | - | |
| GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1) | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OXIDATIVE PHOSPHORYLATION | 135 | 73 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG PARKINSONS DISEASE | 133 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| REACTOME TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT | 141 | 85 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOCHONDRIAL PROTEIN IMPORT | 58 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | 98 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | 16 | 9 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| APPIERTO RESPONSE TO FENRETINIDE DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS UP | 201 | 127 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION DN | 206 | 136 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| MOOTHA VOXPHOS | 87 | 51 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR DN | 25 | 15 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE DN | 87 | 59 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRACX UP | 20 | 14 | All SZGR 2.0 genes in this pathway |
| MARIADASON RESPONSE TO BUTYRATE SULINDAC 6 | 52 | 32 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1B TARGETS | 92 | 68 | All SZGR 2.0 genes in this pathway |
| LEIN LOCALIZED TO PROXIMAL DENDRITES | 37 | 26 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER T7 | 98 | 63 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| MIKHAYLOVA OXIDATIVE STRESS RESPONSE VIA VHL UP | 7 | 6 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |