Gene Page: PARK2
Summary ?
| GeneID | 5071 |
| Symbol | PARK2 |
| Synonyms | AR-JP|LPRS2|PDJ|PRKN |
| Description | parkin RBR E3 ubiquitin protein ligase |
| Reference | MIM:602544|HGNC:HGNC:8607|Ensembl:ENSG00000185345|HPRD:03967|Vega:OTTHUMG00000015970 |
| Gene type | protein-coding |
| Map location | 6q25.2-q27 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias,schizotypal | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 1.0504 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03742724 | 6 | 161838997 | PARK2 | 4.901E-4 | 0.372 | 0.047 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
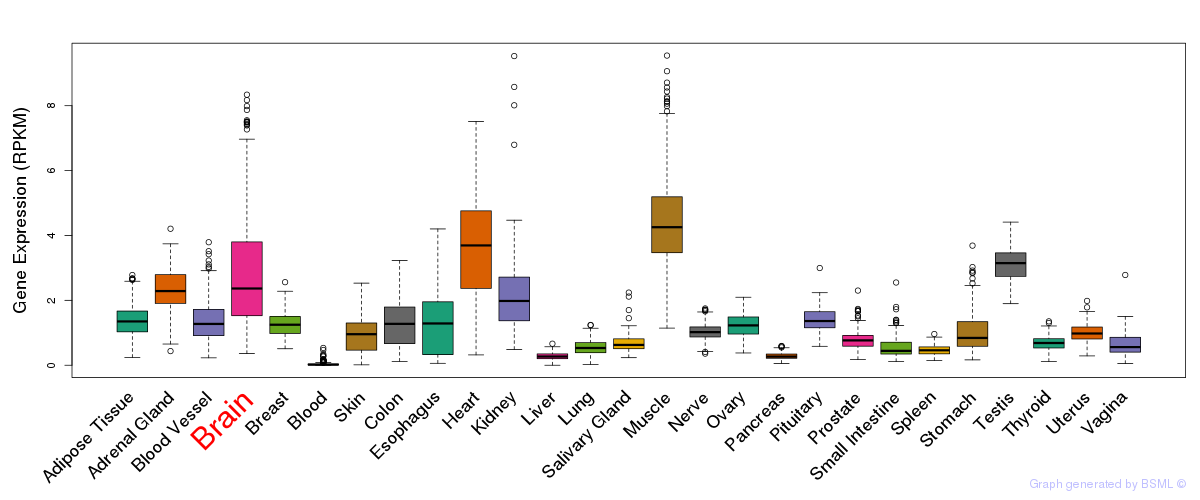
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 11590439 |12364339 |16352719 |17097639 | |
| GO:0004842 | ubiquitin-protein ligase activity | IDA | 17097639 | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0016881 | acid-amino acid ligase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | TAS | Brain (GO term level: 6) | 9560156 |
| GO:0006464 | protein modification process | IEA | - | |
| GO:0042787 | protein ubiquitination during ubiquitin-dependent protein catabolic process | IDA | 17097639 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IDA | 17097639 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0048471 | perinuclear region of cytoplasm | IDA | 17097639 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAG5 | BAG-5 | BCL2-associated athanogene 5 | BAG5 interacts with parkin. | BIND | 15603737 |
| CAMK2A | CAMKA | KIAA0968 | calcium/calmodulin-dependent protein kinase II alpha | Affinity Capture-Western | BioGRID | 11679592 |
| CASK | CAGH39 | CMG | FLJ22219 | FLJ31914 | LIN2 | MICPCH | TNRC8 | calcium/calmodulin-dependent serine protein kinase (MAGUK family) | - | HPRD,BioGRID | 11679592 |
| CASP1 | ICE | IL1BC | P45 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | - | HPRD | 12692130 |
| CCNA1 | - | cyclin A1 | Affinity Capture-Western | BioGRID | 12628165 |
| CCNE1 | CCNE | cyclin E1 | Affinity Capture-Western | BioGRID | 12628165 |
| CUL1 | MGC149834 | MGC149835 | cullin 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12628165 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | Affinity Capture-Western | BioGRID | 11679592 |
| FBXW7 | AGO | CDC4 | DKFZp686F23254 | FBW6 | FBW7 | FBX30 | FBXO30 | FBXW6 | FLJ16457 | SEL-10 | SEL10 | F-box and WD repeat domain containing 7 | - | HPRD,BioGRID | 12628165 |
| GPR37 | EDNRBL | PAELR | hET(B)R-LP | G protein-coupled receptor 37 (endothelin receptor type B-like) | - | HPRD,BioGRID | 11439185 |12150907 |
| GRIN2B | MGC142178 | MGC142180 | NMDAR2B | NR2B | hNR3 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | - | HPRD,BioGRID | 11679592 |
| HOMER1 | HOMER | HOMER1A | HOMER1B | HOMER1C | SYN47 | Ves-1 | homer homolog 1 (Drosophila) | Affinity Capture-Western | BioGRID | 11679592 |
| JTV1 | AIMP2 | P38 | PRO0992 | JTV1 gene | - | HPRD,BioGRID | 12783850 |
| NDUFA4L2 | FLJ26118 | NUOMS | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 | Two-hybrid | BioGRID | 16169070 |
| PAFAH1B2 | - | platelet-activating factor acetylhydrolase, isoform Ib, beta subunit 30kDa | Two-hybrid | BioGRID | 16169070 |
| RAC1 | MGC111543 | MIG5 | TC-25 | p21-Rac1 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | Two-hybrid | BioGRID | 16169070 |
| RAD1 | HRAD1 | REC1 | RAD1 homolog (S. pombe) | Two-hybrid | BioGRID | 16169070 |
| RGS2 | G0S8 | regulator of G-protein signaling 2, 24kDa | Two-hybrid | BioGRID | 16169070 |
| SEPT4 | ARTS | BRADEION | CE5B3 | H5 | MART | PNUTL2 | SEP4 | hCDCREL-2 | hucep-7 | septin 4 | - | HPRD | 14559152 |
| SEPT5 | CDCREL | CDCREL-1 | CDCREL1 | H5 | PNUTL1 | septin 5 | - | HPRD,BioGRID | 11078524 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD,BioGRID | 11588587 |
| SNCAIP | MGC39814 | SYPH1 | synuclein, alpha interacting protein | - | HPRD,BioGRID | 11590439 |
| SNCAIP | MGC39814 | SYPH1 | synuclein, alpha interacting protein | Synphilin-1 is ubiquitinated by Parkin. | BIND | 15105460 |
| SNCAIP | MGC39814 | SYPH1 | synuclein, alpha interacting protein | Synphilin interacts with parkin. | BIND | 15603737 |
| SYT11 | DKFZp781D015 | KIAA0080 | MGC10881 | MGC17226 | SYT12 | synaptotagmin XI | - | HPRD,BioGRID | 12925569 |
| TRIP13 | 16E1BP | thyroid hormone receptor interactor 13 | Two-hybrid | BioGRID | 16189514 |
| UBB | FLJ25987 | MGC8385 | ubiquitin B | Parkin is S-nitrosylated. | BIND | 15105460 |
| UBE2G1 | E217K | UBC7 | UBE2G | ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) | - | HPRD,BioGRID | 11439185 |
| UBE2J1 | CGI-76 | HSPC153 | HSPC205 | HSU93243 | MGC12555 | NCUBE1 | Ubc6p | ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast) | Affinity Capture-Western | BioGRID | 11439185 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | - | HPRD,BioGRID | 11078524 |11278816 |
| UBE2L6 | MGC40331 | RIG-B | UBCH8 | ubiquitin-conjugating enzyme E2L 6 | - | HPRD,BioGRID | 11078524 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| KEGG PARKINSONS DISEASE | 133 | 78 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PARKIN PATHWAY | 13 | 8 | All SZGR 2.0 genes in this pathway |
| PID ALPHA SYNUCLEIN PATHWAY | 33 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| TCGA GLIOBLASTOMA COPY NUMBER DN | 31 | 21 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER DN | 41 | 34 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER UP | 92 | 45 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| TAVAZOIE METASTASIS | 108 | 68 | All SZGR 2.0 genes in this pathway |