Gene Page: PIN1
Summary ?
| GeneID | 5300 |
| Symbol | PIN1 |
| Synonyms | DOD|UBL5 |
| Description | peptidylprolyl cis/trans isomerase, NIMA-interacting 1 |
| Reference | MIM:601052|HGNC:HGNC:8988|Ensembl:ENSG00000127445|HPRD:03031|Vega:OTTHUMG00000180388 |
| Gene type | protein-coding |
| Map location | 19p13 |
| Pascal p-value | 0.228 |
| Sherlock p-value | 0.23 |
| Fetal beta | -1.568 |
| DMG | 2 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0573 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04699474 | 19 | 9945708 | PIN1 | 2.24E-5 | -0.307 | 0.017 | DMG:Wockner_2014 |
| cg22728790 | 19 | 9945653 | PIN1 | 1.08E-7 | -0.007 | 2.34E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
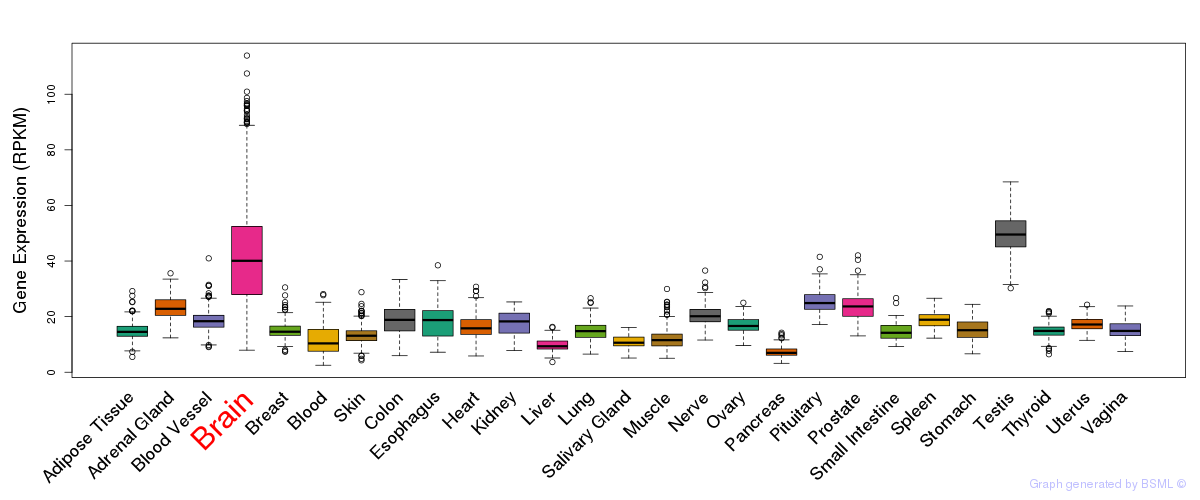
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PTPN3 | 0.87 | 0.59 |
| PTPN4 | 0.86 | 0.81 |
| AKAP2 | 0.85 | 0.74 |
| MCF2 | 0.85 | 0.62 |
| FAM19A4 | 0.84 | 0.33 |
| RORA | 0.84 | 0.47 |
| EPN3 | 0.84 | 0.45 |
| RASSF3 | 0.84 | 0.49 |
| CHRNA3 | 0.83 | 0.53 |
| SLC17A6 | 0.83 | 0.69 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C16orf74 | -0.32 | -0.53 |
| RGS20 | -0.32 | -0.48 |
| C1orf61 | -0.31 | -0.61 |
| CXCL14 | -0.31 | -0.48 |
| OAF | -0.31 | -0.47 |
| CSAG1 | -0.30 | -0.50 |
| CLDN10 | -0.29 | -0.47 |
| AF347015.21 | -0.29 | -0.54 |
| EIF4EBP3 | -0.29 | -0.53 |
| VAMP5 | -0.29 | -0.54 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003755 | peptidyl-prolyl cis-trans isomerase activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 11470801 | |
| GO:0016853 | isomerase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006457 | protein folding | IEA | - | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007088 | regulation of mitosis | TAS | 10391244 | |
| GO:0042127 | regulation of cell proliferation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | TAS | 8606777 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD | 11988841 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | Affinity Capture-Western | BioGRID | 11326318 |
| CCNB1 | CCNB | cyclin B1 | - | HPRD | 9499405 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | PIN1 interacts with Cyclin D1. This interaction was modelled on a demonstrated interaction between human PIN1 and Cyclin D1 from an unspecified species. | BIND | 11805292 |
| CDC2 | CDC28A | CDK1 | DKFZp686L20222 | MGC111195 | cell division cycle 2, G1 to S and G2 to M | - | HPRD | 11774038 |
| CDC25C | CDC25 | cell division cycle 25 homolog C (S. pombe) | - | HPRD | 9482729 |9499405 |10037602|9499405 |10037602 |
| CDC25C | CDC25 | cell division cycle 25 homolog C (S. pombe) | - | HPRD,BioGRID | 9499405 |10037602 |
| CDC27 | APC3 | CDC27Hs | D0S1430E | D17S978E | HNUC | cell division cycle 27 homolog (S. cerevisiae) | - | HPRD,BioGRID | 9499405 |10939594 |
| CDC27 | APC3 | CDC27Hs | D0S1430E | D17S978E | HNUC | cell division cycle 27 homolog (S. cerevisiae) | Pin1 interacts with Cdc27. | BIND | 9499405 |
| CDK9 | C-2k | CDC2L4 | CTK1 | PITALRE | TAK | cyclin-dependent kinase 9 | - | HPRD | 11575923 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD,BioGRID | 11940573 |
| CSNK2A2 | CK2A2 | CSNK2A1 | FLJ43934 | casein kinase 2, alpha prime polypeptide | - | HPRD,BioGRID | 11940573 |
| CSNK2B | CK2B | CK2N | CSK2B | G5A | MGC138222 | MGC138224 | casein kinase 2, beta polypeptide | - | HPRD | 11940573 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD,BioGRID | 11533658 |
| DAB2 | DOC-2 | DOC2 | FLJ26626 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | - | HPRD,BioGRID | 12881709 |
| DDX24 | - | DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 | Two-hybrid | BioGRID | 16169070 |
| GGA2 | FLJ20966 | KIAA1080 | VEAR | golgi associated, gamma adaptin ear containing, ARF binding protein 2 | Two-hybrid | BioGRID | 16189514 |
| GPAA1 | GAA1 | hGAA1 | glycosylphosphatidylinositol anchor attachment protein 1 homolog (yeast) | Two-hybrid | BioGRID | 16169070 |
| HNRNPC | C1 | C2 | HNRNP | HNRPC | MGC104306 | MGC105117 | MGC117353 | MGC131677 | SNRPC | heterogeneous nuclear ribonucleoprotein C (C1/C2) | Two-hybrid | BioGRID | 16189514 |
| JUN | AP-1 | AP1 | c-Jun | jun oncogene | - | HPRD,BioGRID | 11432833 |
| KIF20B | DKFZp434B0435 | DKFZp434P0810 | KRMP1 | MPHOSPH1 | MPP-1 | MPP1 | kinesin family member 20B | - | HPRD,BioGRID | 11470801 |
| KLHL20 | KHLHX | KLEIP | KLHLX | RP3-383J4.3 | kelch-like 20 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| LEPR | CD295 | OBR | leptin receptor | Two-hybrid | BioGRID | 16169070 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | - | HPRD,BioGRID | 10391244 |
| MDFI | I-MF | I-mfa | MyoD family inhibitor | Two-hybrid | BioGRID | 16189514 |
| MYT1 | C20orf36 | MTF1 | MYTI | PLPB1 | myelin transcription factor 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10504341 |
| NFATC2 | KIAA0611 | NFAT1 | NFATP | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | - | HPRD,BioGRID | 11356192 |
| NONO | NMT55 | NRB54 | P54 | P54NRB | non-POU domain containing, octamer-binding | Two-hybrid | BioGRID | 16189514 |
| PKMYT1 | DKFZp547K1610 | FLJ20093 | MYT1 | protein kinase, membrane associated tyrosine/threonine 1 | - | HPRD,BioGRID | 9499405 |10504341 |
| PKMYT1 | DKFZp547K1610 | FLJ20093 | MYT1 | protein kinase, membrane associated tyrosine/threonine 1 | Pin1 interacts with phosphorylated Myt1. | BIND | 10504341 |
| PLK1 | PLK | STPK13 | polo-like kinase 1 (Drosophila) | - | HPRD,BioGRID | 9499405 |
| PLK1 | PLK | STPK13 | polo-like kinase 1 (Drosophila) | Pin1 interacts with PLK1. | BIND | 9499405 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | - | HPRD | 10932246 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Affinity Capture-Western | BioGRID | 10393805 |
| PTOV1 | ACID2 | DKFZp586I111 | MGC71475 | prostate tumor overexpressed 1 | Two-hybrid | BioGRID | 16169070 |
| PTPN1 | PTP1B | protein tyrosine phosphatase, non-receptor type 1 | - | HPRD | 9499405 |
| RAB4A | HRES-1/RAB4 | RAB4 | RAB4A, member RAS oncogene family | - | HPRD,BioGRID | 10888662 |
| RAF1 | CRAF | NS5 | Raf-1 | c-Raf | v-raf-1 murine leukemia viral oncogene homolog 1 | Raf-1 interacts with Pin1. This interaction was modelled on a demonstrated interaction between human Raf-1 and Pin1 from monkey and unspecified species. | BIND | 15664191 |
| RAI1 | DKFZp434A139 | KIAA1820 | MGC12824 | SMCR | SMS | retinoic acid induced 1 | Two-hybrid | BioGRID | 16189514 |
| RBBP8 | CTIP | RIM | retinoblastoma binding protein 8 | Two-hybrid | BioGRID | 16189514 |
| RBPMS | HERMES | RNA binding protein with multiple splicing | Two-hybrid | BioGRID | 16189514 |
| SFRS12 | DKFZp564B176 | MGC133045 | SRrp508 | SRrp86 | splicing factor, arginine/serine-rich 12 | - | HPRD | 14559993 |
| SOCS3 | ATOD4 | CIS3 | Cish3 | MGC71791 | SOCS-3 | SSI-3 | SSI3 | suppressor of cytokine signaling 3 | Two-hybrid | BioGRID | 16169070 |
| SUPT5H | FLJ34157 | SPT5 | SPT5H | suppressor of Ty 5 homolog (S. cerevisiae) | - | HPRD,BioGRID | 11575923 |
| TOP2A | TOP2 | TP2A | topoisomerase (DNA) II alpha 170kDa | - | HPRD,BioGRID | 11940573 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 12388558 |12397362 |
| TP73 | P73 | tumor protein p73 | Pin1 interacts with phosphorylated p73-alpha | BIND | 15175157 |
| TP73 | P73 | tumor protein p73 | Pin1 interacts with p73-beta. | BIND | 15175157 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Two-hybrid | BioGRID | 16189514 |
| TSC22D4 | THG-1 | THG1 | TSC22 domain family, member 4 | Two-hybrid | BioGRID | 16189514 |
| VHL | HRCA1 | RCA1 | VHL1 | von Hippel-Lindau tumor suppressor | Affinity Capture-MS | BioGRID | 17353931 |
| WEE1 | DKFZp686I18166 | FLJ16446 | WEE1A | WEE1hu | WEE1 homolog (S. pombe) | Affinity Capture-Western Far Western | BioGRID | 9499405 |
| ZCCHC10 | FLJ20094 | zinc finger, CCHC domain containing 10 | Two-hybrid | BioGRID | 16169070 |
| ZMIZ2 | DKFZp761I2123 | KIAA1886 | ZIMP7 | hZIMP7 | zinc finger, MIZ-type containing 2 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MPR PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| ST WNT BETA CATENIN PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| PID P73PATHWAY | 79 | 59 | All SZGR 2.0 genes in this pathway |
| PID MYC PATHWAY | 25 | 22 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | 31 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS DN | 260 | 143 | All SZGR 2.0 genes in this pathway |
| BREUHAHN GROWTH FACTOR SIGNALING IN LIVER CANCER | 22 | 19 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| MACLACHLAN BRCA1 TARGETS DN | 16 | 12 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA DN | 52 | 33 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOGONIA | 24 | 17 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS DURATION CORR DN | 146 | 90 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-200bc/429 | 111 | 117 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.