Gene Page: IFT57
Summary ?
| GeneID | 55081 |
| Symbol | IFT57 |
| Synonyms | ESRRBL1|HIPPI|MHS4R2 |
| Description | intraflagellar transport 57 |
| Reference | MIM:606621|HGNC:HGNC:17367|Ensembl:ENSG00000114446|HPRD:09430|Vega:OTTHUMG00000159223 |
| Gene type | protein-coding |
| Map location | 3q13.13 |
| Pascal p-value | 4.378E-4 |
| Sherlock p-value | 1.549E-5 |
| Fetal beta | -0.943 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04359 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.04047 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17232450 | chr3 | 107869894 | IFT57 | 55081 | 0.02 | cis | ||
| rs17232556 | chr3 | 107877462 | IFT57 | 55081 | 0.02 | cis | ||
| rs17232563 | chr3 | 107877537 | IFT57 | 55081 | 0.02 | cis | ||
| rs11928715 | chr3 | 107890217 | IFT57 | 55081 | 0.02 | cis | ||
| rs17829213 | chr3 | 107890926 | IFT57 | 55081 | 0.02 | cis | ||
| rs7632036 | chr3 | 107895325 | IFT57 | 55081 | 0.02 | cis | ||
| rs9840611 | chr3 | 107895760 | IFT57 | 55081 | 0.02 | cis | ||
| rs2701061 | chr3 | 107926125 | IFT57 | 55081 | 0.03 | cis | ||
| rs1289745 | chr3 | 107926257 | IFT57 | 55081 | 0 | cis | ||
| rs1289744 | chr3 | 107926339 | IFT57 | 55081 | 0.01 | cis | ||
| rs11924195 | chr3 | 107926517 | IFT57 | 55081 | 0.02 | cis | ||
| rs1034496 | chr3 | 107939783 | IFT57 | 55081 | 0.03 | cis | ||
| rs1289745 | chr3 | 107926257 | IFT57 | 55081 | 0.19 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
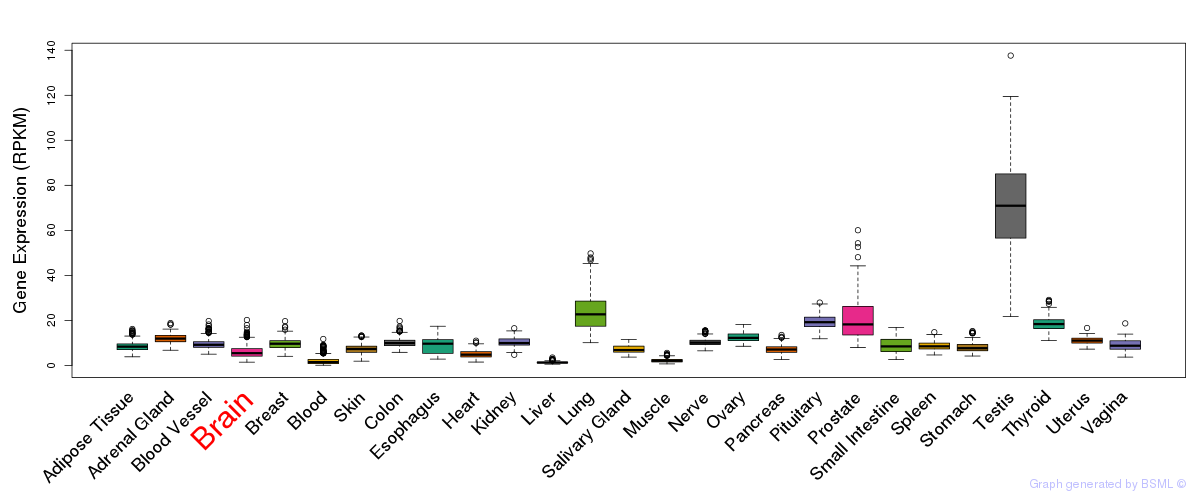
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 18188704 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0006919 | caspase activation | IDA | 11788820 | |
| GO:0042981 | regulation of apoptosis | IDA | 11788820 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005932 | basal body | ISS | - | |
| GO:0005929 | cilium | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER ADVANCED VS EARLY DN | 138 | 70 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 6 7WK DN | 79 | 54 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI DN | 172 | 107 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN T LYMPHOCYTE UP | 12 | 7 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE UP | 156 | 92 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |