Gene Page: AMBRA1
Summary ?
| GeneID | 55626 |
| Symbol | AMBRA1 |
| Synonyms | DCAF3|WDR94 |
| Description | autophagy/beclin-1 regulator 1 |
| Reference | MIM:611359|HGNC:HGNC:25990|Ensembl:ENSG00000110497|HPRD:10975|Vega:OTTHUMG00000166500 |
| Gene type | protein-coding |
| Map location | 11p11.2 |
| Pascal p-value | 5.791E-10 |
| Sherlock p-value | 0.475 |
| Fetal beta | -0.489 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17142575 | chr6 | 6790864 | AMBRA1 | 55626 | 0.17 | trans | ||
| rs1568778 | chr7 | 109250931 | AMBRA1 | 55626 | 0.2 | trans | ||
| rs711457 | chr7 | 109264996 | AMBRA1 | 55626 | 0.08 | trans | ||
| rs16877935 | chr8 | 31216487 | AMBRA1 | 55626 | 0.14 | trans | ||
| rs16877941 | chr8 | 31219122 | AMBRA1 | 55626 | 0.14 | trans | ||
| rs737377 | chr8 | 31248987 | AMBRA1 | 55626 | 0.07 | trans | ||
| rs16877965 | chr8 | 31252303 | AMBRA1 | 55626 | 0.08 | trans | ||
| rs11235411 | chr11 | 71688034 | AMBRA1 | 55626 | 0.04 | trans | ||
| rs9914334 | chr17 | 69795668 | AMBRA1 | 55626 | 0.09 | trans |
Section II. Transcriptome annotation
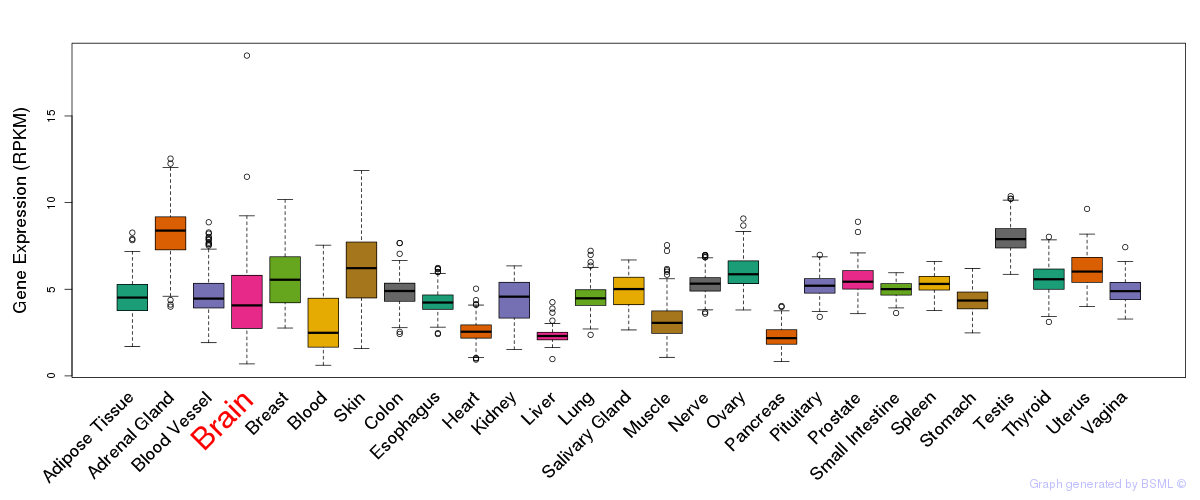
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DNAJC12 | 0.66 | 0.72 |
| VWA5A | 0.65 | 0.71 |
| ADK | 0.65 | 0.70 |
| SCOC | 0.64 | 0.68 |
| MAOA | 0.63 | 0.65 |
| BCHE | 0.63 | 0.60 |
| PDHB | 0.63 | 0.68 |
| CCNE1 | 0.63 | 0.67 |
| SCP2 | 0.63 | 0.65 |
| PAM | 0.63 | 0.70 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SCUBE1 | -0.47 | -0.39 |
| SATB2 | -0.45 | -0.28 |
| NHSL1 | -0.45 | -0.28 |
| SLA | -0.44 | -0.17 |
| TTC28 | -0.43 | -0.21 |
| WDR86 | -0.42 | -0.41 |
| MPPED1 | -0.42 | -0.26 |
| ADRA2A | -0.42 | -0.22 |
| GJC1 | -0.41 | -0.25 |
| NAV2 | -0.41 | -0.21 |
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0006914 | autophagy | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IDA | 17589504 | |
| GO:0031410 | cytoplasmic vesicle | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| DARWICHE SKIN TUMOR PROMOTER DN | 185 | 115 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK LOW DN | 165 | 107 | All SZGR 2.0 genes in this pathway |
| DARWICHE PAPILLOMA RISK HIGH DN | 180 | 110 | All SZGR 2.0 genes in this pathway |
| DARWICHE SQUAMOUS CELL CARCINOMA DN | 181 | 107 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| MIZUSHIMA AUTOPHAGOSOME FORMATION | 19 | 15 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-200bc/429 | 739 | 746 | 1A,m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-23 | 914 | 920 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-323 | 914 | 920 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-485-3p | 907 | 913 | 1A | hsa-miR-485-3p | GUCAUACACGGCUCUCCUCUCU |
| miR-7 | 350 | 357 | 1A,m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
| miR-9 | 999 | 1006 | 1A,m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.