Gene Page: PCDHB11
Summary ?
| GeneID | 56125 |
| Symbol | PCDHB11 |
| Synonyms | ME2|PCDH-BETA11 |
| Description | protocadherin beta 11 |
| Reference | MIM:606337|HGNC:HGNC:8682|HPRD:06955| |
| Gene type | protein-coding |
| Map location | 5q31 |
| Pascal p-value | 0.054 |
| Sherlock p-value | 0.526 |
| Fetal beta | 1.78 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
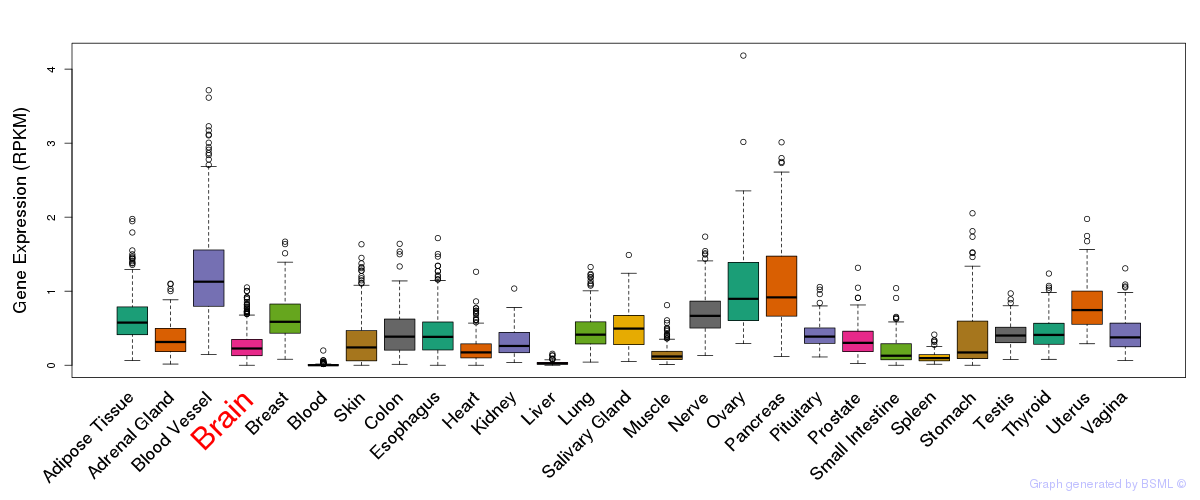
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RHOT2 | 0.89 | 0.88 |
| PPAN-P2RY11 | 0.89 | 0.89 |
| CDK9 | 0.87 | 0.87 |
| ZGPAT | 0.86 | 0.87 |
| SH2B1 | 0.86 | 0.88 |
| C19orf6 | 0.86 | 0.86 |
| WDR24 | 0.85 | 0.83 |
| ACAP3 | 0.85 | 0.87 |
| TUBGCP6 | 0.85 | 0.88 |
| PPAN | 0.85 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.68 | -0.69 |
| AF347015.31 | -0.65 | -0.62 |
| AF347015.27 | -0.63 | -0.62 |
| MT-CO2 | -0.62 | -0.60 |
| AF347015.8 | -0.61 | -0.59 |
| GNG11 | -0.59 | -0.63 |
| MT-CYB | -0.59 | -0.57 |
| AF347015.33 | -0.58 | -0.56 |
| MT-ATP8 | -0.58 | -0.65 |
| NOSTRIN | -0.58 | -0.56 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | NAS | 11322959 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007416 | synaptogenesis | TAS | Synap (GO term level: 6) | 12231349 |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 12231349 |
| GO:0007155 | cell adhesion | TAS | 10380929 | |
| GO:0007156 | homophilic cell adhesion | IEA | - | |
| GO:0016339 | calcium-dependent cell-cell adhesion | NAS | 9182820 |11322959 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | NAS | 9182820 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 10380929 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS HCP WITH H3 UNMETHYLATED | 80 | 50 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES HCP WITH H3K27ME3 | 41 | 30 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 3 UP | 170 | 97 | All SZGR 2.0 genes in this pathway |