Gene Page: RGL2
Summary ?
| GeneID | 5863 |
| Symbol | RGL2 |
| Synonyms | HKE1.5|KE1.5|RAB2L |
| Description | ral guanine nucleotide dissociation stimulator like 2 |
| Reference | MIM:602306|HGNC:HGNC:9769|Ensembl:ENSG00000237441|HPRD:03810|Vega:OTTHUMG00000031072 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 5.614E-6 |
| Sherlock p-value | 0.798 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RGL2 | chr6 | 33264213 | C | T | NM_001243738 NM_004761 | p.68D>N p.150D>N | missense missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
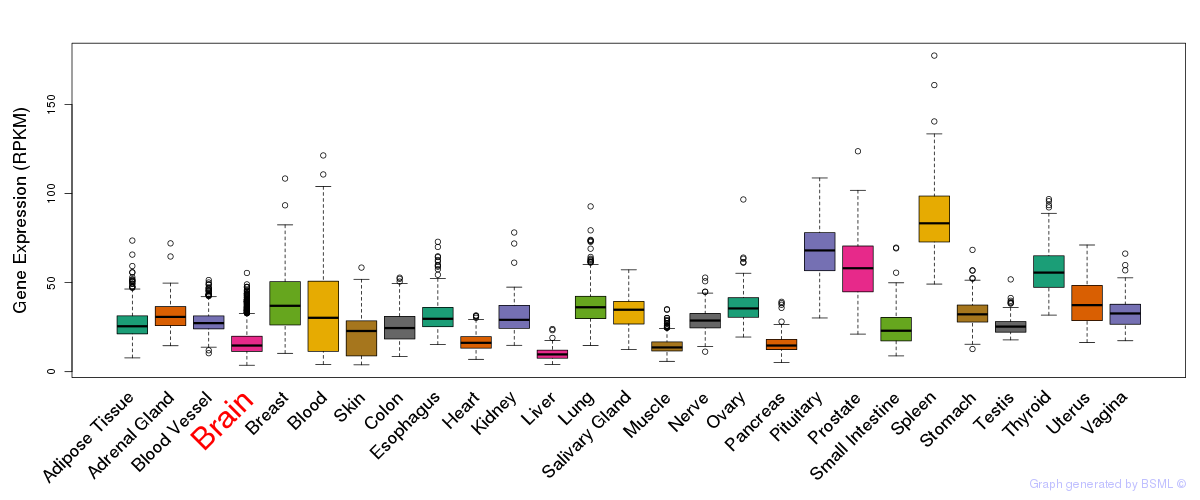
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005085 | guanyl-nucleotide exchange factor activity | IEA | - | |
| GO:0005085 | guanyl-nucleotide exchange factor activity | NAS | - | |
| GO:0005088 | Ras guanyl-nucleotide exchange factor activity | NAS | 8976381 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007265 | Ras protein signal transduction | NAS | 8976381 | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0007264 | small GTPase mediated signal transduction | IEA | - | |
| GO:0051056 | regulation of small GTPase mediated signal transduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005622 | intracellular | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| BOGNI TREATMENT RELATED MYELOID LEUKEMIA DN | 33 | 19 | All SZGR 2.0 genes in this pathway |
| CREIGHTON AKT1 SIGNALING VIA MTOR DN | 23 | 16 | All SZGR 2.0 genes in this pathway |
| FRIDMAN SENESCENCE UP | 77 | 60 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| YU MYC TARGETS DN | 55 | 38 | All SZGR 2.0 genes in this pathway |
| MA MYELOID DIFFERENTIATION UP | 39 | 29 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 DN | 43 | 31 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER DN | 116 | 83 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 401 | 408 | 1A,m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-142-3p | 432 | 438 | m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-196 | 434 | 440 | m8 | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-24 | 324 | 330 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-27 | 402 | 408 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.