Gene Page: BCL2L2
Summary ?
| GeneID | 599 |
| Symbol | BCL2L2 |
| Synonyms | BCL-W|BCL2-L-2|BCLW|PPP1R51 |
| Description | BCL2 like 2 |
| Reference | MIM:601931|HGNC:HGNC:995|Ensembl:ENSG00000129473|HPRD:03569|Vega:OTTHUMG00000028738 |
| Gene type | protein-coding |
| Map location | 14q11.2-q12 |
| Sherlock p-value | 0.198 |
| DEG p-value | DEG:Sanders_2014:DS1_p=0.178:DS1_beta=0.004880:DS2_p=3.17e-05:DS2_beta=0.207:DS2_FDR=1.69e-03 |
| Fetal beta | -2.754 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.047 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
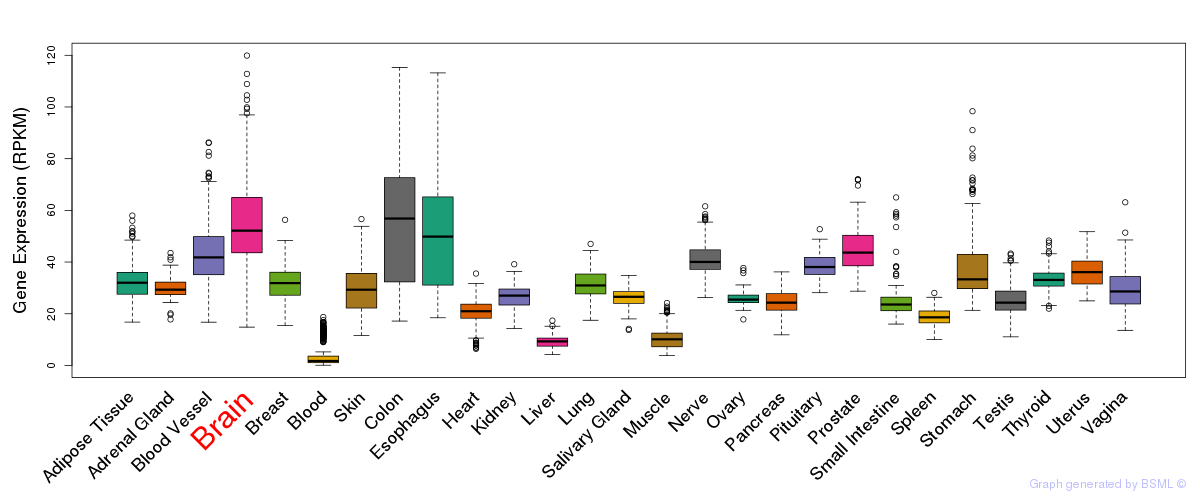
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCAM | 0.60 | 0.72 |
| PERP | 0.59 | 0.68 |
| ZFP36 | 0.59 | 0.49 |
| SSH3 | 0.55 | 0.63 |
| ICAM1 | 0.54 | 0.58 |
| LASS1 | 0.54 | 0.61 |
| SLCO4A1 | 0.52 | 0.57 |
| FOXF1 | 0.50 | 0.51 |
| APOLD1 | 0.49 | 0.36 |
| MOBKL2C | 0.49 | 0.56 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AKT3 | -0.34 | -0.45 |
| ZNF33B | -0.33 | -0.45 |
| PHF14 | -0.33 | -0.52 |
| NLGN1 | -0.33 | -0.40 |
| ROBO2 | -0.32 | -0.44 |
| EPHA7 | -0.32 | -0.44 |
| DOK6 | -0.32 | -0.41 |
| ENY2 | -0.32 | -0.40 |
| TRDMT1 | -0.32 | -0.39 |
| FANCM | -0.32 | -0.42 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 15694340 |16697956 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007283 | spermatogenesis | TAS | 8761287 | |
| GO:0006916 | anti-apoptosis | TAS | 9500547 | |
| GO:0042981 | regulation of apoptosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | NAS | - | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0031966 | mitochondrial membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | - | HPRD,BioGRID | 11483855 |12115603 |
| BAD | BBC2 | BCL2L8 | BCL2-associated agonist of cell death | Bad interacts with Bcl-w. | BIND | 15694340 |
| BAK1 | BAK | BAK-LIKE | BCL2L7 | CDN1 | MGC117255 | MGC3887 | BCL2-antagonist/killer 1 | Affinity Capture-Western | BioGRID | 10381646 |
| BAX | BCL2L4 | BCL2-associated X protein | Affinity Capture-Western | BioGRID | 10381646 |
| BBC3 | JFY1 | PUMA | BCL2 binding component 3 | Puma interacts with Bcl-w. | BIND | 15694340 |
| BCL2L11 | BAM | BIM | BIM-alpha6 | BIM-beta6 | BIM-beta7 | BOD | BimEL | BimL | BCL2-like 11 (apoptosis facilitator) | Bim interacts with Bcl-w. | BIND | 15694340 |
| BCL2L11 | BAM | BIM | BIM-alpha6 | BIM-beta6 | BIM-beta7 | BOD | BimEL | BimL | BCL2-like 11 (apoptosis facilitator) | Two-hybrid | BioGRID | 9731710 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Protein-peptide | BioGRID | 15694340 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Bid interacts with Bcl-w. | BIND | 15694340 |
| BIK | BIP1 | BP4 | NBK | BCL2-interacting killer (apoptosis-inducing) | Affinity Capture-Western | BioGRID | 10381646 |
| BIK | BIP1 | BP4 | NBK | BCL2-interacting killer (apoptosis-inducing) | Bik interacts with Bcl-w. | BIND | 15694340 |
| BMF | FLJ00065 | Bcl2 modifying factor | Bmf interacts with Bcl-w. This interaction was modelled on a demonstrated interaction between mouse Bmf and human Bcl-w. | BIND | 15694340 |
| BMF | FLJ00065 | Bcl2 modifying factor | - | HPRD,BioGRID | 11546872 |
| HRK | DP5 | HARAKIRI | harakiri, BCL2 interacting protein (contains only BH3 domain) | Hrk interacts with Bcl-w. | BIND | 15694340 |
| HRK | DP5 | HARAKIRI | harakiri, BCL2 interacting protein (contains only BH3 domain) | Protein-peptide | BioGRID | 15694340 |
| PMAIP1 | APR | NOXA | phorbol-12-myristate-13-acetate-induced protein 1 | Protein-peptide | BioGRID | 15694340 |
| PPP1CA | MGC15877 | MGC1674 | PP-1A | PPP1A | protein phosphatase 1, catalytic subunit, alpha isoform | Affinity Capture-Western Reconstituted Complex | BioGRID | 12115603 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION DN | 84 | 54 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PREVENT MITOCHONDIAL PERMEABILIZATION | 22 | 16 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| KREPPEL CD99 TARGETS DN | 8 | 5 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| TENEDINI MEGAKARYOCYTE MARKERS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| EHLERS ANEUPLOIDY UP | 41 | 29 | All SZGR 2.0 genes in this pathway |
| MATZUK EMBRYONIC GERM CELL | 19 | 16 | All SZGR 2.0 genes in this pathway |
| MATZUK MALE REPRODUCTION SERTOLI | 28 | 23 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOCYTE | 72 | 55 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATID DIFFERENTIATION | 37 | 26 | All SZGR 2.0 genes in this pathway |
| BONCI TARGETS OF MIR15A AND MIR16 1 | 91 | 75 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| BEIER GLIOMA STEM CELL DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN MYELOMA VS MATURE B LYMPHOCYTE | 101 | 76 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-10 | 1902 | 1908 | 1A | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-103/107 | 1364 | 1370 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-122 | 1331 | 1337 | 1A | hsa-miR-122a | UGGAGUGUGACAAUGGUGUUUGU |
| miR-133 | 1544 | 1551 | 1A,m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-140 | 1954 | 1960 | m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-148/152 | 1450 | 1456 | 1A | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-149 | 1350 | 1357 | 1A,m8 | hsa-miR-149brain | UCUGGCUCCGUGUCUUCACUCC |
| miR-15/16/195/424/497 | 1390 | 1396 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG | ||||
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-17-5p/20/93.mr/106/519.d | 1599 | 1605 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-214 | 1935 | 1941 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-24 | 1348 | 1355 | 1A,m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-29 | 1379 | 1385 | 1A | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-30-3p | 1938 | 1944 | 1A | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.