Gene Page: HCN2
Summary ?
| GeneID | 610 |
| Symbol | HCN2 |
| Synonyms | BCNG-2|BCNG2|HAC-1 |
| Description | hyperpolarization activated cyclic nucleotide gated potassium channel 2 |
| Reference | MIM:602781|HGNC:HGNC:4846|Ensembl:ENSG00000099822|HPRD:07213|Vega:OTTHUMG00000180590 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.117 |
| Sherlock p-value | 0.066 |
| Fetal beta | -0.937 |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
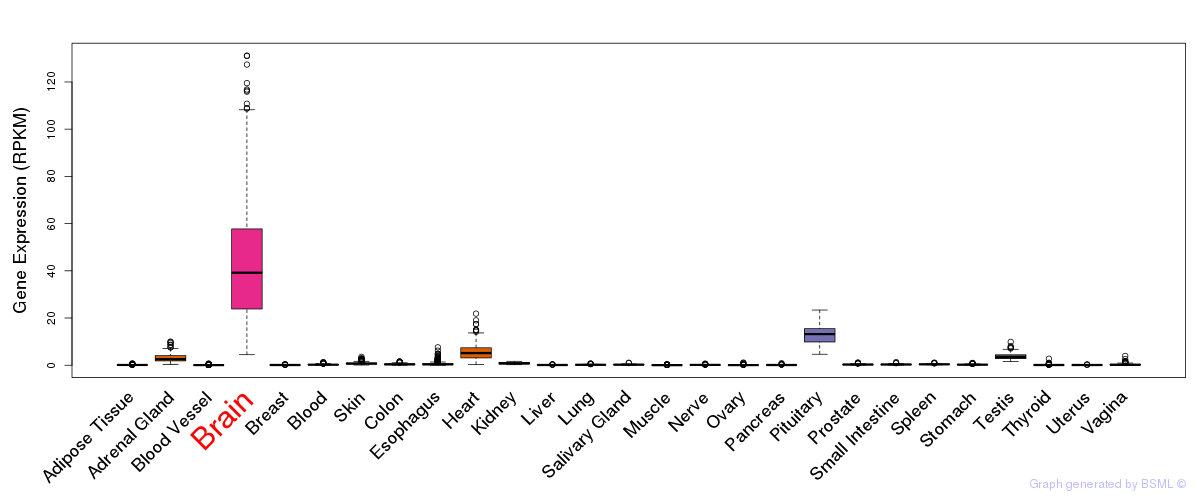
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM171A1 | 0.89 | 0.88 |
| ULK1 | 0.87 | 0.89 |
| SRF | 0.85 | 0.91 |
| AKAP1 | 0.84 | 0.86 |
| MESDC1 | 0.83 | 0.84 |
| RAPGEF1 | 0.83 | 0.89 |
| WDTC1 | 0.82 | 0.88 |
| RUNDC1 | 0.82 | 0.86 |
| DAB2IP | 0.82 | 0.88 |
| FAM168B | 0.82 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.68 | -0.76 |
| AF347015.31 | -0.63 | -0.67 |
| MT-CO2 | -0.62 | -0.66 |
| C1orf54 | -0.62 | -0.74 |
| GNG11 | -0.60 | -0.67 |
| SYCP3 | -0.60 | -0.68 |
| AF347015.8 | -0.59 | -0.64 |
| HIGD1B | -0.58 | -0.65 |
| NOSTRIN | -0.58 | -0.63 |
| MYL3 | -0.57 | -0.62 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0005249 | voltage-gated potassium channel activity | IEA | - | |
| GO:0005272 | sodium channel activity | IEA | - | |
| GO:0030552 | cAMP binding | IEA | - | |
| GO:0030955 | potassium ion binding | IEA | - | |
| GO:0031402 | sodium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007267 | cell-cell signaling | TAS | 9630217 | |
| GO:0006814 | sodium ion transport | IEA | - | |
| GO:0006812 | cation transport | TAS | 10228147 | |
| GO:0006813 | potassium ion transport | IEA | - | |
| GO:0006936 | muscle contraction | TAS | 10228147 | |
| GO:0042391 | regulation of membrane potential | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0008076 | voltage-gated potassium channel complex | TAS | 9630217 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME POTASSIUM CHANNELS | 98 | 68 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| MCCLUNG DELTA FOSB TARGETS 8WK | 47 | 38 | All SZGR 2.0 genes in this pathway |
| LEIN OLIGODENDROCYTE MARKERS | 74 | 53 | All SZGR 2.0 genes in this pathway |
| LEIN PONS MARKERS | 89 | 59 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-135 | 60 | 66 | m8 | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA | ||||
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-181 | 580 | 587 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-25/32/92/363/367 | 391 | 398 | 1A,m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-9 | 56 | 62 | m8 | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.