Gene Page: S100A12
Summary ?
| GeneID | 6283 |
| Symbol | S100A12 |
| Synonyms | CAAF1|CAGC|CGRP|ENRAGE|MRP-6|MRP6|p6 |
| Description | S100 calcium binding protein A12 |
| Reference | MIM:603112|HGNC:HGNC:10489|Ensembl:ENSG00000163221|HPRD:04383|Vega:OTTHUMG00000013127 |
| Gene type | protein-coding |
| Map location | 1q21 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs529819 | chr1 | 14531406 | S100A12 | 6283 | 0.07 | trans | ||
| rs16982680 | chr2 | 16885880 | S100A12 | 6283 | 0.09 | trans | ||
| rs1882577 | chr2 | 227574206 | S100A12 | 6283 | 0.01 | trans | ||
| rs2228048 | chr3 | 30713841 | S100A12 | 6283 | 0.01 | trans | ||
| rs3773648 | chr3 | 30716287 | S100A12 | 6283 | 0.04 | trans | ||
| rs17026432 | chr3 | 30770741 | S100A12 | 6283 | 0.14 | trans | ||
| rs17026436 | chr3 | 30771248 | S100A12 | 6283 | 0.14 | trans | ||
| rs6599081 | chr3 | 40273499 | S100A12 | 6283 | 0.05 | trans | ||
| rs6549200 | chr3 | 69373073 | S100A12 | 6283 | 0.06 | trans | ||
| rs11128125 | chr3 | 69389613 | S100A12 | 6283 | 0.03 | trans | ||
| rs11915229 | chr3 | 140758709 | S100A12 | 6283 | 0.19 | trans | ||
| rs16860219 | chr3 | 185493589 | S100A12 | 6283 | 0.16 | trans | ||
| rs17021660 | chr4 | 147543413 | S100A12 | 6283 | 0.13 | trans | ||
| rs1891150 | chr6 | 128763428 | S100A12 | 6283 | 0.1 | trans | ||
| rs1341597 | chr6 | 128779157 | S100A12 | 6283 | 0.2 | trans | ||
| rs6933496 | chr6 | 129046844 | S100A12 | 6283 | 0.18 | trans | ||
| rs11980248 | chr7 | 97315272 | S100A12 | 6283 | 0.12 | trans | ||
| rs2916688 | chr8 | 6221560 | S100A12 | 6283 | 0.12 | trans | ||
| rs4909503 | chr8 | 136671520 | S100A12 | 6283 | 0.19 | trans | ||
| rs10124001 | chr9 | 4976185 | S100A12 | 6283 | 0.2 | trans | ||
| rs11144447 | chr9 | 78111361 | S100A12 | 6283 | 0.11 | trans | ||
| rs7039109 | chr9 | 109716110 | S100A12 | 6283 | 0 | trans | ||
| rs11202852 | chr10 | 90544072 | S100A12 | 6283 | 0.14 | trans | ||
| rs7948100 | chr11 | 123895762 | S100A12 | 6283 | 0.01 | trans | ||
| rs213141 | chr17 | 32830469 | S100A12 | 6283 | 0.05 | trans | ||
| rs6095518 | chr20 | 48048870 | S100A12 | 6283 | 0.05 | trans | ||
| rs17003861 | chrX | 101204311 | S100A12 | 6283 | 0.01 | trans |
Section II. Transcriptome annotation
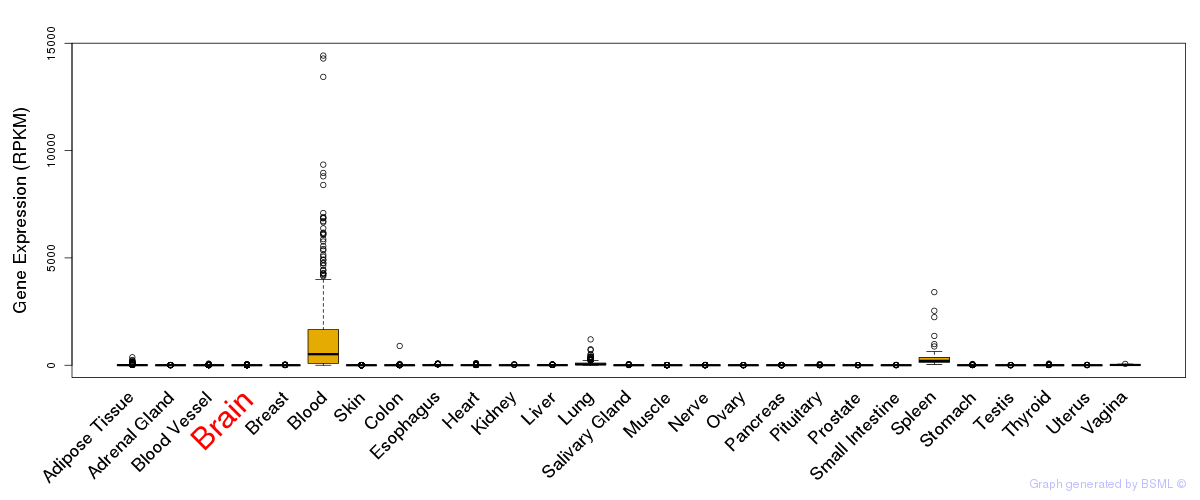
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | TAS | 7626002 | |
| GO:0008270 | zinc ion binding | TAS | 11522286 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006805 | xenobiotic metabolic process | IDA | 11522286 | |
| GO:0006954 | inflammatory response | TAS | 8619876 | |
| GO:0042742 | defense response to bacterium | IEA | - | |
| GO:0050832 | defense response to fungus | IDA | 11522286 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | TAS | 7626002 | |
| GO:0005626 | insoluble fraction | TAS | 7626002 | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005886 | plasma membrane | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRIF MEDIATED TLR3 SIGNALING | 74 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | 23 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED NFKB ACTIVATION | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | 77 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB AND MAP KINASES ACTIVATION MEDIATED BY TLR4 SIGNALING REPERTOIRE | 72 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 8D DN | 205 | 127 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS UP | 149 | 84 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN | 101 | 66 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER C | 113 | 47 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO MERCAPTOPURINE AND LD MTX DN | 21 | 11 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION DN | 128 | 90 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA DN | 41 | 27 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| AUJLA IL22 AND IL17A SIGNALING | 11 | 5 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA DN | 68 | 45 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |