Gene Page: MRPL44
Summary ?
| GeneID | 65080 |
| Symbol | MRPL44 |
| Synonyms | COXPD16|L44MT|MRP-L44 |
| Description | mitochondrial ribosomal protein L44 |
| Reference | MIM:611849|HGNC:HGNC:16650|Ensembl:ENSG00000135900|HPRD:14763|Vega:OTTHUMG00000133164 |
| Gene type | protein-coding |
| Map location | 2q36.1 |
| Pascal p-value | 0.348 |
| Sherlock p-value | 0.323 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00916 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01016 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01766534 | 2 | 224822249 | MRPL44 | 1.72E-8 | -0.007 | 6.25E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
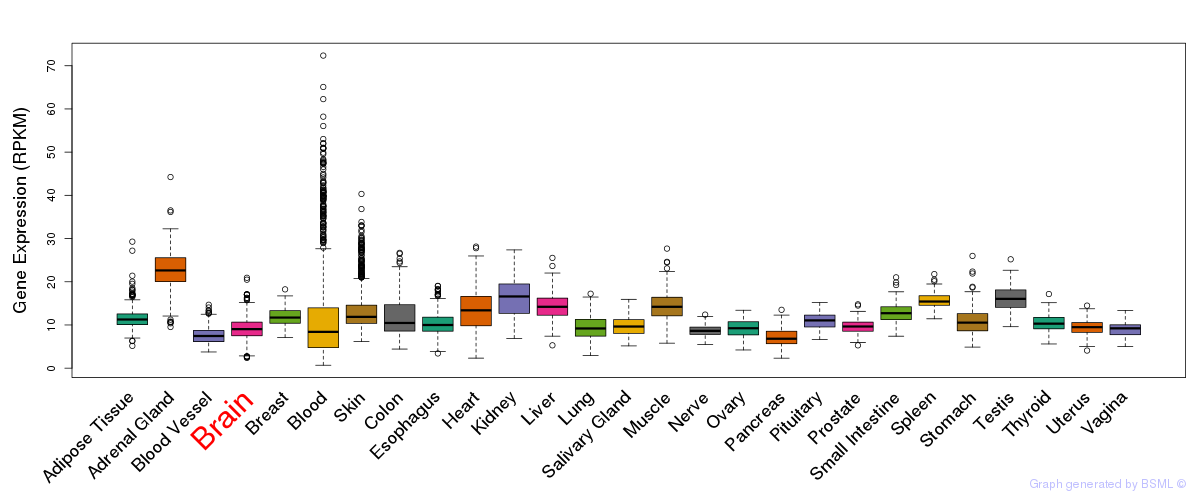
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| GO:0003725 | double-stranded RNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0004519 | endonuclease activity | IEA | - | |
| GO:0004525 | ribonuclease III activity | IEA | - | |
| GO:0016787 | hydrolase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006396 | RNA processing | IEA | - | |
| GO:0008150 | biological_process | ND | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005840 | ribosome | IEA | - | |
| GO:0005575 | cellular_component | ND | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005739 | mitochondrion | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TURASHVILI BREAST DUCTAL CARCINOMA VS LOBULAR NORMAL UP | 73 | 50 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL CIS | 65 | 38 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |