Gene Page: SNAP25
Summary ?
| GeneID | 6616 |
| Symbol | SNAP25 |
| Synonyms | CMS18|RIC-4|RIC4|SEC9|SNAP|SNAP-25|bA416N4.2|dJ1068F16.2 |
| Description | synaptosome associated protein 25kDa |
| Reference | MIM:600322|HGNC:HGNC:11132|Ensembl:ENSG00000132639|HPRD:02637|Vega:OTTHUMG00000031863 |
| Gene type | protein-coding |
| Map location | 20p12-p11.2 |
| Pascal p-value | 0.083 |
| Sherlock p-value | 0.651 |
| Fetal beta | -3.912 |
| DMG | 1 (# studies) |
| Support | CANABINOID DOPAMINE EXOCYTOSIS METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.046 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 8 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0523 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06433879 | 20 | 10200138 | SNAP25 | 5.923E-4 | -0.448 | 0.05 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
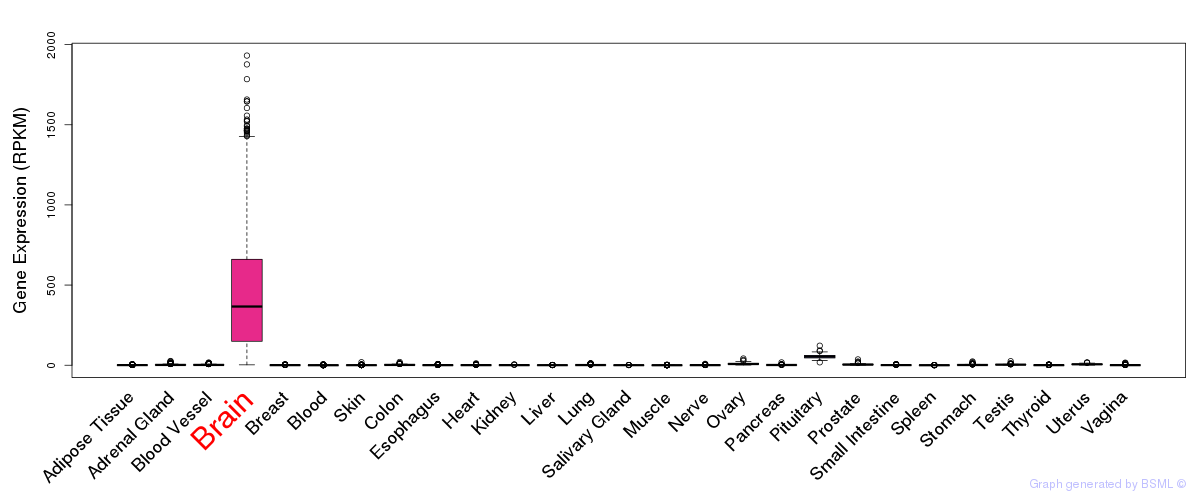
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ARX | 0.93 | 0.84 |
| DLX2 | 0.88 | 0.82 |
| DLX1 | 0.87 | 0.77 |
| E2F2 | 0.85 | 0.70 |
| DLX5 | 0.85 | 0.76 |
| SPC24 | 0.85 | 0.62 |
| SP9 | 0.84 | 0.83 |
| AC018470.3 | 0.84 | 0.83 |
| CDCA7 | 0.84 | 0.74 |
| HJURP | 0.84 | 0.67 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.36 | -0.65 |
| HLA-F | -0.34 | -0.56 |
| AF347015.31 | -0.34 | -0.68 |
| S100B | -0.34 | -0.63 |
| AF347015.27 | -0.34 | -0.66 |
| MT-CO2 | -0.33 | -0.69 |
| AF347015.33 | -0.33 | -0.66 |
| PTGDS | -0.33 | -0.56 |
| CA4 | -0.33 | -0.55 |
| TINAGL1 | -0.33 | -0.52 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 8692999 |11438518 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007269 | neurotransmitter secretion | NAS | neuron, axon, Synap, serotonin, Neurotransmitter, dopamine (GO term level: 8) | - |
| GO:0007268 | synaptic transmission | NAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 8760387 |
| GO:0016081 | synaptic vesicle docking during exocytosis | NAS | Synap (GO term level: 10) | 8760387 |
| GO:0001504 | neurotransmitter uptake | NAS | neuron, Cholinergic, Synap, Neurotransmitter, Glial (GO term level: 8) | 8760387 |
| GO:0050796 | regulation of insulin secretion | TAS | 15537656 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043005 | neuron projection | ISS | neuron, axon, neurite, dendrite (GO term level: 5) | - |
| GO:0031201 | SNARE complex | ISS | neuron, Synap (GO term level: 6) | - |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0030426 | growth cone | ISS | axon, dendrite (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0016020 | membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CPLX1 | CPX-I | CPX1 | complexin 1 | Co-crystal Structure Reconstituted Complex | BioGRID | 11832227 |12200427 |
| DNM1 | DNM | dynamin 1 | - | HPRD | 10373452 |
| GOSR1 | GOLIM2 | GOS-28 | GOS28 | GOS28/P28 | GS28 | P28 | golgi SNAP receptor complex member 1 | Reconstituted Complex | BioGRID | 9325254 |
| HGS | HRS | ZFYVE8 | hepatocyte growth factor-regulated tyrosine kinase substrate | - | HPRD | 10825299 |11493665|11493665 |
| HGS | HRS | ZFYVE8 | hepatocyte growth factor-regulated tyrosine kinase substrate | - | HPRD | 11493665 |
| ITSN1 | ITSN | MGC134948 | MGC134949 | SH3D1A | SH3P17 | intersectin 1 (SH3 domain protein) | - | HPRD,BioGRID | 10373452 |
| KCNB1 | DRK1 | KV2.1 | h-DRK1 | potassium voltage-gated channel, Shab-related subfamily, member 1 | - | HPRD,BioGRID | 12403834 |
| KIF5B | KINH | KNS | KNS1 | UKHC | kinesin family member 5B | - | HPRD,BioGRID | 12475239 |
| NAPA | SNAPA | N-ethylmaleimide-sensitive factor attachment protein, alpha | Reconstituted Complex | BioGRID | 7622514 |
| PRRT2 | DKFZp547J199 | FLJ25513 | proline-rich transmembrane protein 2 | Two-hybrid | BioGRID | 16169070 |
| SCAMP1 | SCAMP | SCAMP37 | secretory carrier membrane protein 1 | Reconstituted Complex | BioGRID | 10777571 |
| SNAPIN | SNAPAP | SNAP-associated protein | - | HPRD,BioGRID | 10195194 |
| SNAPIN | SNAPAP | SNAP-associated protein | SNAP25 interacts with Snapin. | BIND | 15635093 |
| SNIP | KIAA1684 | SNAP25-interacting protein | - | HPRD,BioGRID | 10625663 |
| STX11 | FHL4 | HLH4 | HPLH4 | syntaxin 11 | Two-hybrid | BioGRID | 16169070 |16189514 |
| STX12 | MGC51957 | STX13 | STX14 | syntaxin 12 | - | HPRD,BioGRID | 10886332 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | - | HPRD | 7622514 |7768895 |9556632 |10373452 |11509230|10954418 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | - | HPRD | 7622514 |9556632|10954418 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | Affinity Capture-Western Co-crystal Structure Reconstituted Complex Two-hybrid | BioGRID | 7553862 |7768895 |7961655 |8663154 |9852078 |10195194 |10449403 |10954418 |11524423 |11832227 |16169070 |
| STX2 | EPIM | EPM | MGC51014 | STX2A | STX2B | STX2C | syntaxin 2 | - | HPRD,BioGRID | 7768895 |
| STX3 | STX3A | syntaxin 3 | - | HPRD,BioGRID | 7768895 |
| STX4 | STX4A | p35-2 | syntaxin 4 | - | HPRD | 8760387 |
| STX4 | STX4A | p35-2 | syntaxin 4 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 7768895 |8663154 |9852078 |10194441 |
| STX5 | SED5 | STX5A | syntaxin 5 | Reconstituted Complex | BioGRID | 9325254 |
| STXBP1 | EIEE4 | MUNC18-1 | UNC18 | hUNC18 | rbSec1 | syntaxin binding protein 1 | - | HPRD | 9395480 |
| STXBP6 | FLJ39638 | HSPC156 | amisyn | syntaxin binding protein 6 (amisyn) | - | HPRD | 12145319 |
| SYT1 | DKFZp781D2042 | P65 | SVP65 | SYT | synaptotagmin I | - | HPRD,BioGRID | 10692432 |
| SYT3 | DKFZp761O132 | SytIII | synaptotagmin III | - | HPRD,BioGRID | 10692432 |
| SYT9 | FLJ45896 | synaptotagmin IX | - | HPRD | 12062043 |
| TRIM9 | KIAA0282 | RNF91 | SPRING | tripartite motif-containing 9 | - | HPRD,BioGRID | 11524423 |
| UNC13B | MGC133279 | MGC133280 | MUNC13 | UNC13 | Unc13h2 | hmunc13 | unc-13 homolog B (C. elegans) | Co-fractionation | BioGRID | 8999968 |
| VAMP2 | FLJ11460 | SYB2 | VAMP-2 | vesicle-associated membrane protein 2 (synaptobrevin 2) | - | HPRD | 7961655 |
| VAMP2 | FLJ11460 | SYB2 | VAMP-2 | vesicle-associated membrane protein 2 (synaptobrevin 2) | Affinity Capture-Western Co-crystal Structure Far Western Reconstituted Complex Two-hybrid | BioGRID | 9030619 |11524423 |11832227 |
| VAMP7 | SYBL1 | TI-VAMP | VAMP-7 | vesicle-associated membrane protein 7 | - | HPRD,BioGRID | 12853575 |
| VAMP8 | EDB | vesicle-associated membrane protein 8 (endobrevin) | - | HPRD | 10336434 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SNARE INTERACTIONS IN VESICULAR TRANSPORT | 38 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME BOTULINUM NEUROTOXICITY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RELEASE CYCLE | 34 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | 11 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | 10 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN SYNTHESIS AND PROCESSING | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | 17 | 16 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS UP | 44 | 23 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| WATTEL AUTONOMOUS THYROID ADENOMA UP | 73 | 47 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 20 HELA | 11 | 8 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| NELSON RESPONSE TO ANDROGEN UP | 86 | 61 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| LEIN NEURON MARKERS | 69 | 45 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS DN | 39 | 30 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER HCP WITH H3K27ME3 | 97 | 72 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| VICENT METASTASIS UP | 14 | 11 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 360 | 367 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-128 | 354 | 360 | 1A | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU | ||||
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 498 | 505 | 1A,m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-138 | 692 | 698 | 1A | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-153 | 1022 | 1028 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-200bc/429 | 661 | 667 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC | ||||
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-27 | 1103 | 1109 | m8 | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC | ||||
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-342 | 1105 | 1111 | 1A | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-361 | 1055 | 1061 | 1A | hsa-miR-361brain | UUAUCAGAAUCUCCAGGGGUAC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.