Gene Page: STK11
Summary ?
| GeneID | 6794 |
| Symbol | STK11 |
| Synonyms | LKB1|PJS|hLKB1 |
| Description | serine/threonine kinase 11 |
| Reference | MIM:602216|HGNC:HGNC:11389|Ensembl:ENSG00000118046|HPRD:03740| |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.05 |
| Sherlock p-value | 0.425 |
| Fetal beta | -0.566 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0208 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17267676 | 19 | 1194783 | STK11 | 8.6E-9 | -0.01 | 3.99E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | STK11 | 6794 | 0 | trans | ||
| rs7584986 | chr2 | 184111432 | STK11 | 6794 | 0.02 | trans | ||
| rs7787830 | chr7 | 98797019 | STK11 | 6794 | 0.01 | trans | ||
| rs11216033 | chr11 | 98457483 | STK11 | 6794 | 0.09 | trans | ||
| rs16955618 | chr15 | 29937543 | STK11 | 6794 | 9.798E-5 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
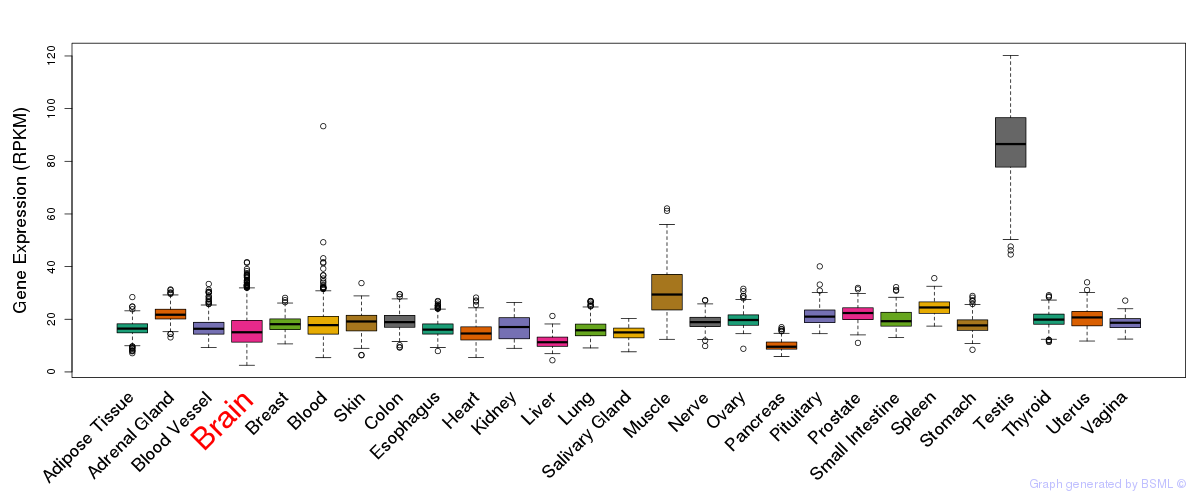
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| COX5B | 0.85 | 0.71 |
| TMEM219 | 0.85 | 0.74 |
| COX4I1 | 0.83 | 0.68 |
| NENF | 0.83 | 0.69 |
| MSRB2 | 0.82 | 0.72 |
| HINT2 | 0.81 | 0.68 |
| SEPW1 | 0.81 | 0.68 |
| C19orf70 | 0.81 | 0.67 |
| C12orf62 | 0.81 | 0.70 |
| NDUFB9 | 0.81 | 0.64 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BAT2D1 | -0.61 | -0.58 |
| MACF1 | -0.55 | -0.53 |
| BDP1 | -0.55 | -0.54 |
| ZC3H13 | -0.54 | -0.53 |
| BOD1L | -0.54 | -0.53 |
| UPF2 | -0.53 | -0.50 |
| CHD2 | -0.53 | -0.53 |
| RAPH1 | -0.52 | -0.51 |
| MDN1 | -0.52 | -0.50 |
| GIGYF2 | -0.52 | -0.49 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0000287 | magnesium ion binding | IDA | 12805220 | |
| GO:0005515 | protein binding | IPI | 17216128 | |
| GO:0005524 | ATP binding | IDA | 12805220 | |
| GO:0004674 | protein serine/threonine kinase activity | IDA | 12805220 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0030145 | manganese ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IDA | 12805220 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007050 | cell cycle arrest | IDA | 12805220 |17216128 | |
| GO:0008285 | negative regulation of cell proliferation | IMP | 17216128 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 12805220 | |
| GO:0005737 | cytoplasm | IDA | 12805220 |17216128 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CAB39 | CGI-66 | FLJ22682 | MO25 | calcium binding protein 39 | Affinity Capture-Western | BioGRID | 15561763 |
| CAB39 | CGI-66 | FLJ22682 | MO25 | calcium binding protein 39 | - | HPRD | 14676191 |
| CDC37 | P50CDC37 | cell division cycle 37 homolog (S. cerevisiae) | - | HPRD,BioGRID | 12489981 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12489981 |
| MARK2 | EMK1 | MGC99619 | PAR-1 | Par1b | MAP/microtubule affinity-regulating kinase 2 | - | HPRD | 14976552 |
| MARK4 | FLJ90097 | KIAA1860 | MARKL1 | Nbla00650 | MAP/microtubule affinity-regulating kinase 4 | LKB1 phosphorylates MARK4. | BIND | 14676191 |
| PRKAA2 | AMPK | AMPK2 | PRKAA | protein kinase, AMP-activated, alpha 2 catalytic subunit | LKB1 interacts with AMPK-alpha. | BIND | 14985505 |
| SMARCA4 | BAF190 | BRG1 | FLJ39786 | SNF2 | SNF2-BETA | SNF2L4 | SNF2LB | SWI2 | hSNF2b | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 | - | HPRD,BioGRID | 11445556 |
| STK11 | LKB1 | PJS | serine/threonine kinase 11 | LKB1 autophosphorylates itself. | BIND | 14676191 |
| STK11IP | KIAA1898 | LIP1 | LKB1IP | STK11IP1 | serine/threonine kinase 11 interacting protein | - | HPRD,BioGRID | 11741830 |
| STRADA | FLJ90524 | LYK5 | NY-BR-96 | PMSE | STRAD | Stlk | STE20-related kinase adaptor alpha | - | HPRD,BioGRID | 12805220 |
| STRADB | ALS2CR2 | CALS-21 | ILPIP | ILPIPA | MGC102916 | PAPK | PRO1038 | STE20-related kinase adaptor beta | LKB1 phosphorylates PAPK. | BIND | 14676191 |
| TNIP2 | ABIN-2 | ABIN2 | FLIP1 | KLIP | MGC4289 | TNFAIP3 interacting protein 2 | - | HPRD | 12595760 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 11430832 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG ADIPOCYTOKINE SIGNALING PATHWAY | 67 | 57 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | 19 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME PKB MEDIATED EVENTS | 29 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE | 71 | 51 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| BIDUS METASTASIS DN | 161 | 93 | All SZGR 2.0 genes in this pathway |
| CORRADETTI MTOR PATHWAY REGULATORS DN | 6 | 5 | All SZGR 2.0 genes in this pathway |
| GALLUZZI PERMEABILIZE MITOCHONDRIA | 43 | 31 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED SIGNIFICANTLY | 26 | 22 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED FREQUENTLY | 12 | 10 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL AND BRAIN QTL TRANS | 185 | 114 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| DING LUNG CANCER MUTATED RECURRENTLY | 6 | 5 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR TARGETS UP | 48 | 33 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |