Gene Page: STXBP1
Summary ?
| GeneID | 6812 |
| Symbol | STXBP1 |
| Synonyms | MUNC18-1|NSEC1|P67|RBSEC1|UNC18 |
| Description | syntaxin binding protein 1 |
| Reference | MIM:602926|HGNC:HGNC:11444|Ensembl:ENSG00000136854|HPRD:04235|Vega:OTTHUMG00000020713 |
| Gene type | protein-coding |
| Map location | 9q34.1 |
| Pascal p-value | 0.342 |
| Sherlock p-value | 0.807 |
| Fetal beta | -1.019 |
| DMG | 1 (# studies) |
| Support | CANABINOID DOPAMINE EXOCYTOSIS METABOTROPIC GLUTAMATE RECEPTOR SEROTONIN G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_mitochondria G2Cdb.human_TAP-PSD-95-CORE G2Cdb.humanARC G2Cdb.humanNRC CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0549 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09332592 | 9 | 130374679 | STXBP1 | 2.15E-8 | -0.009 | 7.29E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
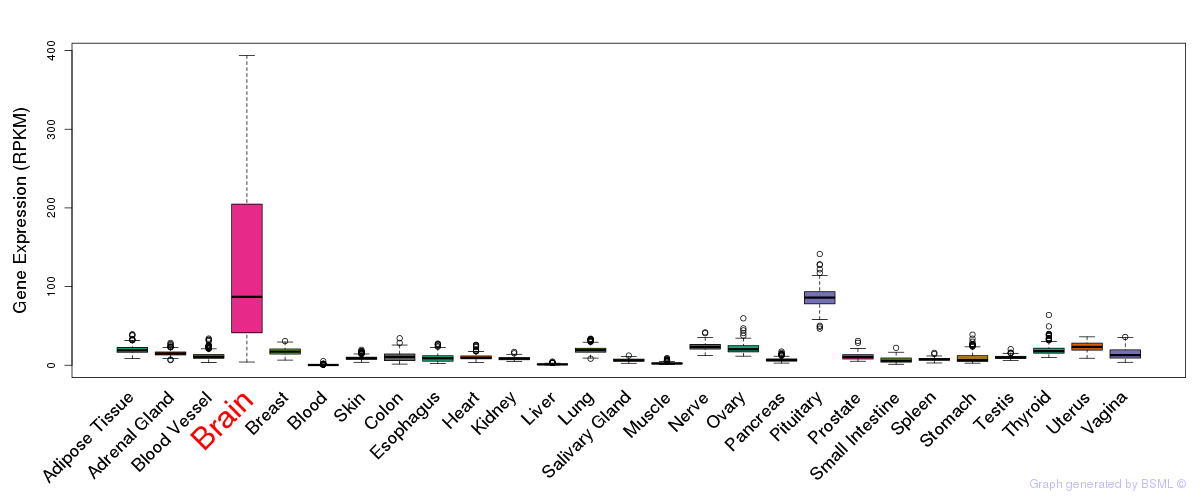
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0006904 | vesicle docking during exocytosis | IEA | - | |
| GO:0016192 | vesicle-mediated transport | IEA | - | |
| GO:0015031 | protein transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016020 | membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APBA1 | D9S411E | MINT1 | X11 | X11A | X11ALPHA | amyloid beta (A4) precursor protein-binding, family A, member 1 | - | HPRD | 9395480 |
| CDK5 | PSSALRE | cyclin-dependent kinase 5 | - | HPRD,BioGRID | 12963086 |
| DOC2A | - | double C2-like domains, alpha | - | HPRD,BioGRID | 9115738 |
| DOC2B | - | double C2-like domains, beta | - | HPRD,BioGRID | 9115738 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | - | HPRD,BioGRID | 12963086 |
| NEFH | NFH | neurofilament, heavy polypeptide | - | HPRD | 12963086 |
| NEFM | NEF3 | NF-M | NFM | neurofilament, medium polypeptide | - | HPRD | 12963086 |
| SNAP25 | FLJ23079 | RIC-4 | RIC4 | SEC9 | SNAP | SNAP-25 | bA416N4.2 | dJ1068F16.2 | synaptosomal-associated protein, 25kDa | - | HPRD | 9395480 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | Affinity Capture-Western Reconstituted Complex | BioGRID | 7553862 |7768895 |10449403 |12093152 |12963086 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | - | HPRD | 9045631 |
| STX1B | STX1B1 | STX1B2 | syntaxin 1B | Reconstituted Complex | BioGRID | 12093152 |
| STX2 | EPIM | EPM | MGC51014 | STX2A | STX2B | STX2C | syntaxin 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 7768895 |12773094 |
| STX2 | EPIM | EPM | MGC51014 | STX2A | STX2B | STX2C | syntaxin 2 | - | HPRD | 9045631 |
| STX3 | STX3A | syntaxin 3 | - | HPRD | 9045631 |
| STX3 | STX3A | syntaxin 3 | Reconstituted Complex | BioGRID | 7768895 |
| STX4 | STX4A | p35-2 | syntaxin 4 | Affinity Capture-Western Reconstituted Complex | BioGRID | 7768895 |12773094 |
| SYTL4 | DKFZp451P0116 | FLJ40960 | SLP4 | synaptotagmin-like 4 | Affinity Capture-Western | BioGRID | 12590134 |
| TUBA4A | FLJ30169 | H2-ALPHA | TUBA1 | tubulin, alpha 4a | Affinity Capture-Western | BioGRID | 12963086 |
| TUBB2A | TUBB | TUBB2 | dJ40E16.7 | tubulin, beta 2A | Affinity Capture-Western | BioGRID | 12963086 |
| USO1 | P115 | TAP | VDP | USO1 homolog, vesicle docking protein (yeast) | - | HPRD | 10903204 |11927603 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RELEASE CYCLE | 34 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | 11 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | 10 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | 17 | 16 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2A DN | 141 | 84 | All SZGR 2.0 genes in this pathway |
| HU ANGIOGENESIS UP | 21 | 16 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 2 | 127 | 92 | All SZGR 2.0 genes in this pathway |
| STARK HYPPOCAMPUS 22Q11 DELETION UP | 53 | 40 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH PML RARA FUSION | 77 | 62 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| FRASOR RESPONSE TO ESTRADIOL DN | 82 | 52 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS POSTNATAL | 63 | 50 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS DN | 39 | 30 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS UP | 112 | 71 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP | 139 | 95 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 12 | 30 | 20 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-134 | 1259 | 1265 | 1A | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-141/200a | 1743 | 1749 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-15/16/195/424/497 | 1225 | 1231 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG | ||||
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-218 | 1761 | 1768 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-410 | 1659 | 1665 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.