Gene Page: VAMP2
Summary ?
| GeneID | 6844 |
| Symbol | VAMP2 |
| Synonyms | SYB2|VAMP-2 |
| Description | vesicle associated membrane protein 2 |
| Reference | MIM:185881|HGNC:HGNC:12643|Ensembl:ENSG00000220205|HPRD:01717|Vega:OTTHUMG00000150254 |
| Gene type | protein-coding |
| Map location | 17p13.1 |
| Pascal p-value | 0.361 |
| Sherlock p-value | 0.801 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=-1.11:CC_BA10_disease_P=0.0403:HBB_BA9_fold_change=-1.28:HBB_BA9_disease_P=0.0379 |
| Fetal beta | -1.939 |
| Support | EXOCYTOSIS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_mGluR5 G2Cdb.human_TAP-PSD-95-CORE CompositeSet Darnell FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
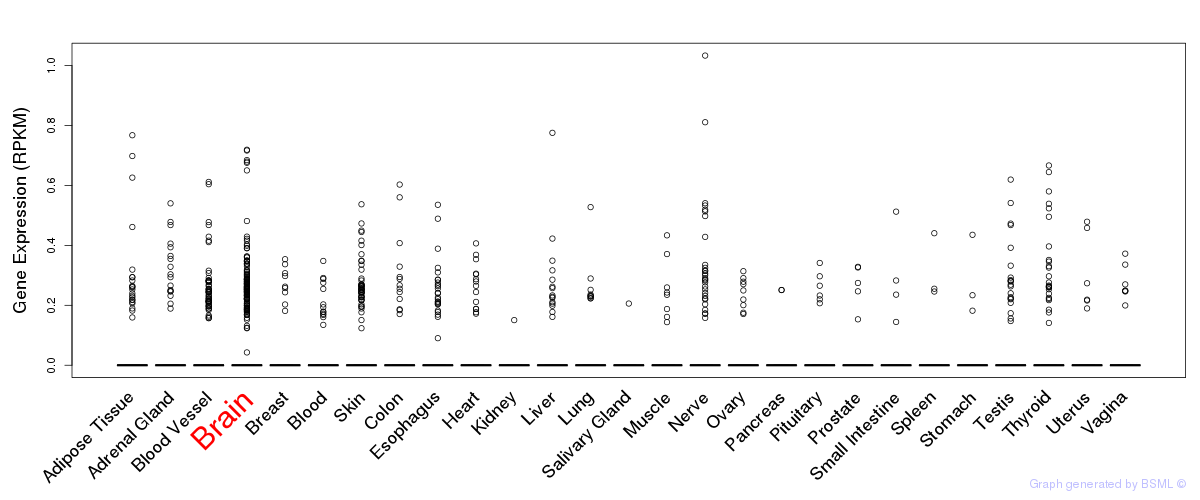
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DNAJC28 | 0.74 | 0.69 |
| FBXO9 | 0.73 | 0.64 |
| TTC35 | 0.69 | 0.67 |
| C2orf74 | 0.69 | 0.61 |
| KRT222P | 0.68 | 0.58 |
| NMNAT1 | 0.68 | 0.70 |
| C13orf1 | 0.68 | 0.59 |
| C1D | 0.67 | 0.60 |
| SGTB | 0.66 | 0.56 |
| TYW3 | 0.66 | 0.57 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HYAL2 | -0.44 | -0.44 |
| OLFM2 | -0.41 | -0.38 |
| SEMA4B | -0.40 | -0.35 |
| SH3BP2 | -0.39 | -0.39 |
| AC006276.1 | -0.39 | -0.34 |
| NAPRT1 | -0.39 | -0.33 |
| SH2B2 | -0.39 | -0.41 |
| SPNS1 | -0.38 | -0.32 |
| SLC38A5 | -0.38 | -0.40 |
| WDR86 | -0.38 | -0.37 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 9920726 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016192 | vesicle-mediated transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 1976629 | |
| GO:0030054 | cell junction | IEA | - | |
| GO:0031410 | cytoplasmic vesicle | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BCAP31 | 6C6-AG | BAP31 | CDM | DXS1357E | B-cell receptor-associated protein 31 | Reconstituted Complex | BioGRID | 9396746 |
| CPLX1 | CPX-I | CPX1 | complexin 1 | - | HPRD,BioGRID | 11832227 |
| DNAJC5 | CSP | DKFZp434N1429 | DKFZp761N1221 | DNAJC5A | FLJ00118 | FLJ13070 | DnaJ (Hsp40) homolog, subfamily C, member 5 | Csp1 interacts with VAMP2. This interaction was modeled on a demonstrated interaction between cow Csp1, and human VAMP2. | BIND | 15610015 |
| NAPA | SNAPA | N-ethylmaleimide-sensitive factor attachment protein, alpha | Reconstituted Complex | BioGRID | 7622514 |
| RABAC1 | PRA1 | PRAF1 | YIP3 | Rab acceptor 1 (prenylated) | - | HPRD,BioGRID | 9341137 |
| SNAP23 | HsT17016 | SNAP23A | SNAP23B | synaptosomal-associated protein, 23kDa | SNAP23 interacts with VAMP2. | BIND | 15610015 |
| SNAP23 | HsT17016 | SNAP23A | SNAP23B | synaptosomal-associated protein, 23kDa | - | HPRD,BioGRID | 10713150 |
| SNAP25 | FLJ23079 | RIC-4 | RIC4 | SEC9 | SNAP | SNAP-25 | bA416N4.2 | dJ1068F16.2 | synaptosomal-associated protein, 25kDa | - | HPRD | 7961655 |
| SNAP25 | FLJ23079 | RIC-4 | RIC4 | SEC9 | SNAP | SNAP-25 | bA416N4.2 | dJ1068F16.2 | synaptosomal-associated protein, 25kDa | Affinity Capture-Western Co-crystal Structure Far Western Reconstituted Complex Two-hybrid | BioGRID | 9030619 |11524423 |11832227 |
| STX11 | FHL4 | HLH4 | HPLH4 | syntaxin 11 | - | HPRD,BioGRID | 10036234 |
| STX1A | HPC-1 | STX1 | p35-1 | syntaxin 1A (brain) | Affinity Capture-Western Co-crystal Structure Far Western Reconstituted Complex Two-hybrid | BioGRID | 7553862 |9030619 |10100611 |10449403 |11832227 |12093152 |
| STX1B | STX1B1 | STX1B2 | syntaxin 1B | - | HPRD,BioGRID | 12093152 |
| STX2 | EPIM | EPM | MGC51014 | STX2A | STX2B | STX2C | syntaxin 2 | Two-hybrid | BioGRID | 12828989 |
| STX3 | STX3A | syntaxin 3 | Two-hybrid | BioGRID | 12828989 |
| STX4 | STX4A | p35-2 | syntaxin 4 | Affinity Capture-Western Two-hybrid | BioGRID | 8760387 |10194441 |10820264 |12517971 |
| STX4 | STX4A | p35-2 | syntaxin 4 | - | HPRD | 8760387 |12417022 |12517971|8770861 |
| STXBP2 | Hunc18b | MUNC18-2 | UNC18-2 | UNC18B | pp10122 | syntaxin binding protein 2 | - | HPRD | 12773094 |
| STXBP3 | MUNC18-3 | MUNC18C | PSP | UNC-18C | syntaxin binding protein 3 | Reconstituted Complex | BioGRID | 12832401 |
| SYPL1 | H-SP1 | SYPL | synaptophysin-like 1 | - | HPRD | 10370136 |
| TRPC3 | TRP3 | transient receptor potential cation channel, subfamily C, member 3 | - | HPRD | 15327778 |
| TRPC3 | TRP3 | transient receptor potential cation channel, subfamily C, member 3 | TRPC3 interacts with VAMP2. | BIND | 15327778 |
| UNC13B | MGC133279 | MGC133280 | MUNC13 | UNC13 | Unc13h2 | hmunc13 | unc-13 homolog B (C. elegans) | Co-fractionation | BioGRID | 8999968 |
| VAMP3 | CEB | vesicle-associated membrane protein 3 (cellubrevin) | Reconstituted Complex | BioGRID | 12130530 |
| VAMP8 | EDB | vesicle-associated membrane protein 8 (endobrevin) | Reconstituted Complex | BioGRID | 12130530 |
| VAPA | MGC3745 | VAP-33 | VAP-A | VAP33 | hVAP-33 | VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa | - | HPRD,BioGRID | 9657962 |
| VAPB | ALS8 | VAMP-B | VAMP-C | VAP-B | VAP-C | VAMP (vesicle-associated membrane protein)-associated protein B and C | - | HPRD,BioGRID | 9920726 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SNARE INTERACTIONS IN VESICULAR TRANSPORT | 38 | 25 | All SZGR 2.0 genes in this pathway |
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| PID INSULIN GLUCOSE PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME MEMBRANE TRAFFICKING | 129 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME BOTULINUM NEUROTOXICITY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | 60 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | 15 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RELEASE CYCLE | 34 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | 11 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME DIABETES PATHWAYS | 133 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | 10 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN SYNTHESIS AND PROCESSING | 21 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME LYSOSOME VESICLE BIOGENESIS | 23 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | 53 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | 17 | 16 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| STARK HYPPOCAMPUS 22Q11 DELETION UP | 53 | 40 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| MODY HIPPOCAMPUS POSTNATAL | 63 | 50 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MCCLUNG COCAINE REWARD 5D | 79 | 62 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| RAMPON ENRICHED LEARNING ENVIRONMENT EARLY UP | 15 | 12 | All SZGR 2.0 genes in this pathway |
| AMBROSINI FLAVOPIRIDOL TREATMENT TP53 | 109 | 63 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 36HR UP | 221 | 150 | All SZGR 2.0 genes in this pathway |
| STEIN ESR1 TARGETS | 85 | 55 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| ZEMBUTSU SENSITIVITY TO DOXORUBICIN | 17 | 10 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 946 | 952 | m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA | ||||
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-127 | 98 | 104 | m8 | hsa-miR-127brain | UCGGAUCCGUCUGAGCUUGGCU |
| miR-133 | 912 | 918 | 1A | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-135 | 70 | 76 | 1A | hsa-miR-135a | UAUGGCUUUUUAUUCCUAUGUGA |
| hsa-miR-135b | UAUGGCUUUUCAUUCCUAUGUG | ||||
| miR-150 | 522 | 528 | m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-153 | 325 | 332 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-25/32/92/363/367 | 328 | 334 | 1A | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-28 | 1234 | 1240 | m8 | hsa-miR-28brain | AAGGAGCUCACAGUCUAUUGAG |
| miR-30-3p | 1185 | 1192 | 1A,m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-325 | 202 | 208 | 1A | hsa-miR-325 | CCUAGUAGGUGUCCAGUAAGUGU |
| miR-338 | 1553 | 1560 | 1A,m8 | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA | ||||
| miR-34/449 | 1226 | 1232 | m8 | hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU |
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU | ||||
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| hsa-miR-34abrain | UGGCAGUGUCUUAGCUGGUUGUU | ||||
| hsa-miR-34c | AGGCAGUGUAGUUAGCUGAUUGC | ||||
| hsa-miR-449 | UGGCAGUGUAUUGUUAGCUGGU | ||||
| hsa-miR-449b | AGGCAGUGUAUUGUUAGCUGGC | ||||
| miR-34b | 1100 | 1106 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-369-3p | 169 | 175 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 169 | 175 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-377 | 1534 | 1540 | 1A | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-409-5p | 1688 | 1694 | 1A | hsa-miR-409-5p | AGGUUACCCGAGCAACUUUGCA |
| miR-410 | 171 | 177 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-448 | 326 | 332 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-491 | 1481 | 1487 | 1A | hsa-miR-491brain | AGUGGGGAACCCUUCCAUGAGGA |
| miR-493-5p | 1666 | 1673 | 1A,m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.