Gene Page: TSC2
Summary ?
| GeneID | 7249 |
| Symbol | TSC2 |
| Synonyms | LAM|PPP1R160|TSC4 |
| Description | tuberous sclerosis 2 |
| Reference | MIM:191092|HGNC:HGNC:12363|Ensembl:ENSG00000103197|HPRD:01850|Vega:OTTHUMG00000128745 |
| Gene type | protein-coding |
| Map location | 16p13.3 |
| Pascal p-value | 0.256 |
| Sherlock p-value | 0.935 |
| Fetal beta | -0.517 |
| eGene | Cerebellar Hemisphere Cerebellum |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0217 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
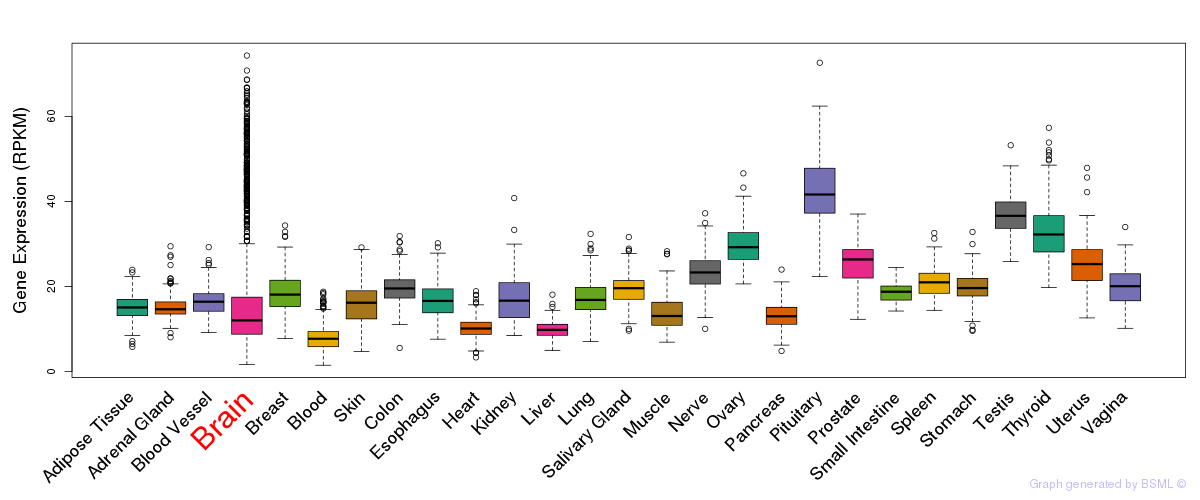
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TTC7B | 0.52 | 0.53 |
| AKTIP | 0.52 | 0.53 |
| AUH | 0.52 | 0.52 |
| STAR | 0.52 | 0.49 |
| EPDR1 | 0.51 | 0.51 |
| SLC25A40 | 0.51 | 0.47 |
| DNAJA4 | 0.51 | 0.50 |
| IDH3A | 0.51 | 0.52 |
| HSPA4L | 0.50 | 0.51 |
| C1orf196 | 0.50 | 0.49 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC010300.1 | -0.27 | -0.24 |
| RP9P | -0.26 | -0.30 |
| BCL7C | -0.25 | -0.31 |
| RBMX2 | -0.25 | -0.22 |
| AC005921.3 | -0.25 | -0.28 |
| FAM159B | -0.24 | -0.35 |
| CLEC3B | -0.23 | -0.29 |
| SH2D2A | -0.23 | -0.23 |
| ANP32C | -0.23 | -0.21 |
| AF347015.18 | -0.23 | -0.08 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005096 | GTPase activator activity | IDA | 9045618 | |
| GO:0005515 | protein binding | IPI | 9580671 |10585443 |12364343 | |
| GO:0042803 | protein homodimerization activity | IPI | 10585443 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001843 | neural tube closure | ISS | - | |
| GO:0030100 | regulation of endocytosis | ISS | - | |
| GO:0006469 | negative regulation of protein kinase activity | ISS | - | |
| GO:0006606 | protein import into nucleus | ISS | - | |
| GO:0008285 | negative regulation of cell proliferation | ISS | - | |
| GO:0014067 | negative regulation of phosphoinositide 3-kinase cascade | ISS | - | |
| GO:0007507 | heart development | ISS | - | |
| GO:0008104 | protein localization | ISS | - | |
| GO:0048009 | insulin-like growth factor receptor signaling pathway | ISS | - | |
| GO:0051056 | regulation of small GTPase mediated signal transduction | IEA | - | |
| GO:0050918 | positive chemotaxis | ISS | - | |
| GO:0045786 | negative regulation of cell cycle | IEA | - | |
| GO:0043547 | positive regulation of GTPase activity | IEA | - | |
| GO:0046626 | regulation of insulin receptor signaling pathway | ISS | - | |
| GO:0051898 | negative regulation of protein kinase B signaling cascade | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IDA | 8806680 | |
| GO:0005794 | Golgi apparatus | IDA | 10585443 | |
| GO:0005829 | cytosol | EXP | 15314020 |15589136 | |
| GO:0005829 | cytosol | IDA | 10585443 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005624 | membrane fraction | IDA | 16636147 | |
| GO:0005634 | nucleus | IDA | 17114346 | |
| GO:0005634 | nucleus | IDA | 16636147 | |
| GO:0005737 | cytoplasm | IDA | 9580671 |16636147 | |
| GO:0005737 | cytoplasm | IDA | 17114346 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0048471 | perinuclear region of cytoplasm | IDA | 8806680 | |
| GO:0033596 | TSC1-TSC2 complex | IDA | 9580671 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | - | HPRD,BioGRID | 12167664 |
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | Akt interacts with and phosphorylates tuberin. This interaction was modelled on a demonstrated interaction between Akt from an unspecified species and human Tuberin. | BIND | 12150915 |
| AXIN1 | AXIN | MGC52315 | axin 1 | Affinity Capture-Western | BioGRID | 12511557 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | Co-purification | BioGRID | 12147258 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | Affinity Capture-Western | BioGRID | 16357142 |
| CCND2 | KIAK0002 | MGC102758 | cyclin D2 | Affinity Capture-Western | BioGRID | 16357142 |
| CCND3 | - | cyclin D3 | Affinity Capture-Western | BioGRID | 16357142 |
| CDKN1B | CDKN4 | KIP1 | MEN1B | MEN4 | P27KIP1 | cyclin-dependent kinase inhibitor 1B (p27, Kip1) | Affinity Capture-Western | BioGRID | 15355997 |
| FOXO1 | FKH1 | FKHR | FOXO1A | forkhead box O1 | Affinity Capture-Western Co-localization Reconstituted Complex | BioGRID | 17077083 |
| GSK3B | - | glycogen synthase kinase 3 beta | Affinity Capture-Western Biochemical Activity | BioGRID | 12511557 |16959574 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 15851026 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | Erk2 interacts with TSC2. This interaction was modelled on a demonstrated interaction between Erk2 from an unspecified species and human TSC2. | BIND | 15851026 |
| NEK1 | DKFZp686D06121 | DKFZp686K12169 | KIAA1901 | MGC138800 | NY-REN-55 | NIMA (never in mitosis gene a)-related kinase 1 | NEK1 interacts with Tuberin. | BIND | 14690447 |
| PAM | PAL | PHM | peptidylglycine alpha-amidating monooxygenase | Affinity Capture-Western Co-localization Two-hybrid | BioGRID | 14559897 |
| PLK1 | PLK | STPK13 | polo-like kinase 1 (Drosophila) | Affinity Capture-Western | BioGRID | 16339216 |
| PRKAA1 | AMPK | AMPKa1 | MGC33776 | MGC57364 | protein kinase, AMP-activated, alpha 1 catalytic subunit | Affinity Capture-Western Reconstituted Complex | BioGRID | 14651849 |15261145 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 17043358 |
| RAB5A | RAB5 | RAB5A, member RAS oncogene family | - | HPRD | 9045618 |
| RAB5A | RAB5 | RAB5A, member RAS oncogene family | Co-purification | BioGRID | 12147258 |
| RABEP1 | RAB5EP | RABPT5 | rabaptin, RAB GTPase binding effector protein 1 | - | HPRD | 9045618 |
| RALA | MGC48949 | RAL | v-ral simian leukemia viral oncogene homolog A (ras related) | Affinity Capture-Western | BioGRID | 12842888 |
| RAP1A | KREV-1 | KREV1 | RAP1 | SMGP21 | RAP1A, member of RAS oncogene family | Affinity Capture-Western Co-purification | BioGRID | 12147258 |12842888 |
| RHEB | MGC111559 | RHEB2 | Ras homolog enriched in brain | - | HPRD,BioGRID | 12842888 |
| RHEB | MGC111559 | RHEB2 | Ras homolog enriched in brain | TSC2 interacts with Rheb. | BIND | 15854902 |
| RPS6KA1 | HU-1 | MAPKAPK1A | RSK | RSK1 | ribosomal protein S6 kinase, 90kDa, polypeptide 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 15342917 |15757502 |
| SFN | YWHAS | stratifin | - | HPRD | 12438239 |
| SKP2 | FBL1 | FBXL1 | FLB1 | MGC1366 | S-phase kinase-associated protein 2 (p45) | Phenotypic Suppression | BioGRID | 15355997 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | Affinity Capture-MS Affinity Capture-Western Co-localization Co-purification Two-hybrid | BioGRID | 9580671 |10585443 |10807585 |10915759 |11175345 |11290735 |11741832 |11741833 |12167664 |12176984 |12468542 |12511557 |14551205 |14651849 |15340059 |15851026 |15963462 |16339216 |16424383 |16636147 |17077083 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | - | HPRD | 10915759 |11741833 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | - | HPRD | 9580671 |10915759 |11741833|10915759 |11741833 |
| TSC1 | KIAA0243 | LAM | MGC86987 | TSC | tuberous sclerosis 1 | Hamartin interacts with tuberin | BIND | 9580671 |
| UBE3A | ANCR | AS | E6-AP | EPVE6AP | FLJ26981 | HPVE6A | ubiquitin protein ligase E3A | Reconstituted Complex | BioGRID | 15175323 |
| YWHAA | - | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, alpha polypeptide | Affinity Capture-Western Two-hybrid | BioGRID | 12176984 |12364343 |12468542 |12582162 |16636147 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | - | HPRD | 12438239 |12468542 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | The tumor suppressor protein tuberin interacts with 14-3-3-beta isoform. This interaction is modeled on demonstrated interaction between human tuberin and mouse 14-3-3-beta isoform. | BIND | 12176984 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | - | HPRD | 12438239 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | Tumor suppressor protein tuberin interacts with 14-3-3-epsilon isoform. This interaction is modeled on demonstrated interaction between human tuberin and mouse 14-3-3-epsilon isoform. | BIND | 12176984 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | - | HPRD | 12438239 |
| YWHAG | 14-3-3GAMMA | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | The tumor suppressor protein tuberin interacts with 14-3-3-gamma isoform. This interaction is modeled on demonstrated interaction between human tuberin and mouse 14-3-3-gamma. | BIND | 12176984 |
| YWHAH | YWHA1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | - | HPRD | 12438239 |
| YWHAH | YWHA1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | The tumor suppressor protein tuberin interacts with 14-3-3-eta isoform. This interaction is modeled on demonstrated interaction between human tuberin and mouse 14-3-3-eta isoform. | BIND | 12176984 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Tumor suppressor protein tuberin interacts with 14-3-3-tau isoform. This interaction is modeled on demonstrated interaction between human tuberin and mouse 14-3-3-tau isoform. | BIND | 12176984 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | - | HPRD | 12438239 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD | 12176984 |12438239 |12468542 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | - | HPRD,BioGRID | 12176984 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Tumor suppressor protein tuberin interacts with 14-3-3-zeta isoform. | BIND | 11533041 |12176984 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VDR PATHWAY | 12 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MTOR PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN CARDIAC MYOCTES | 67 | 54 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| PID MTOR 4PATHWAY | 69 | 55 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID P38 MK2 PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID MYC REPRESS PATHWAY | 63 | 52 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB4 SIGNALING | 38 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K EVENTS IN ERBB2 SIGNALING | 44 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | 97 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME INSULIN RECEPTOR SIGNALLING CASCADE | 87 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K AKT ACTIVATION | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME GAB1 SIGNALOSOME | 38 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME PI 3K CASCADE | 56 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | 18 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME PKB MEDIATED EVENTS | 29 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY INSULIN RECEPTOR | 108 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PIP3 ACTIVATES AKT SIGNALING | 29 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME PI3K CASCADE | 71 | 51 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| APPIERTO RESPONSE TO FENRETINIDE UP | 38 | 26 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 16P13 AMPLICON | 120 | 49 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY HNE AND H2O2 | 39 | 29 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE E2 | 28 | 21 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |