Gene Page: ZYX
Summary ?
| GeneID | 7791 |
| Symbol | ZYX |
| Synonyms | ESP-2|HED-2 |
| Description | zyxin |
| Reference | MIM:602002|HGNC:HGNC:13200|Ensembl:ENSG00000159840|HPRD:03592|Vega:OTTHUMG00000023822 |
| Gene type | protein-coding |
| Map location | 7q32 |
| Pascal p-value | 0.06 |
| Sherlock p-value | 0.528 |
| Fetal beta | -0.294 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.01 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05102190 | 7 | 143078242 | ZYX | 3.84E-5 | -0.23 | 0.02 | DMG:Wockner_2014 |
| cg02471194 | 7 | 143078198 | ZYX | 5.7E-5 | -0.317 | 0.022 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17328164 | chr19 | 34424089 | ZYX | 7791 | 0.11 | trans |
Section II. Transcriptome annotation
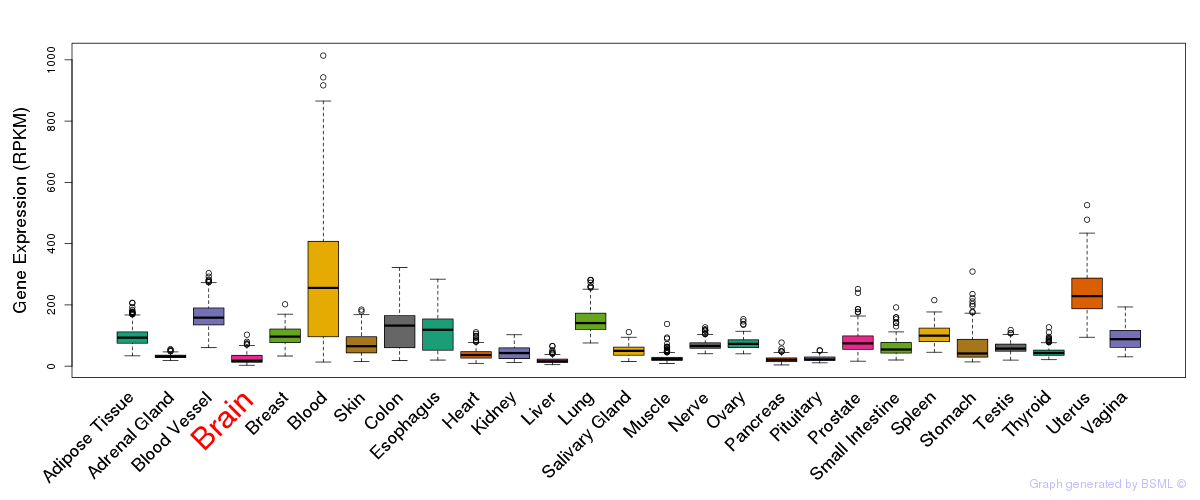
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| WDR27 | 0.91 | 0.88 |
| ANKRD10 | 0.91 | 0.88 |
| FBN3 | 0.90 | 0.92 |
| POLE | 0.90 | 0.93 |
| TRPV1 | 0.90 | 0.85 |
| MYO19 | 0.89 | 0.79 |
| CCNL2 | 0.89 | 0.84 |
| RAD52 | 0.89 | 0.80 |
| RBM5 | 0.89 | 0.89 |
| GAL3ST4 | 0.89 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.57 | -0.69 |
| FBXO2 | -0.57 | -0.65 |
| C5orf53 | -0.56 | -0.76 |
| ACYP2 | -0.56 | -0.66 |
| ALDOC | -0.56 | -0.64 |
| S100B | -0.55 | -0.76 |
| CA4 | -0.55 | -0.72 |
| ACOT13 | -0.55 | -0.70 |
| AIFM3 | -0.55 | -0.65 |
| HLA-C | -0.54 | -0.68 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 10831611 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | TAS | 8940160 | |
| GO:0007267 | cell-cell signaling | TAS | 8940160 | |
| GO:0007165 | signal transduction | TAS | 8940160 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001725 | stress fiber | IDA | 18297730 | |
| GO:0005856 | cytoskeleton | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005925 | focal adhesion | IDA | 18029348 |18297730 | |
| GO:0005913 | cell-cell adherens junction | IDA | 8940160 | |
| GO:0005886 | plasma membrane | TAS | 8940160 | |
| GO:0005887 | integral to plasma membrane | TAS | 8940160 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTN1 | FLJ40884 | actinin, alpha 1 | - | HPRD,BioGRID | 10224105 |11423549 |
| BCAR1 | CAS | CAS1 | CASS1 | CRKAS | P130Cas | breast cancer anti-estrogen resistance 1 | Affinity Capture-Western | BioGRID | 11782456 |
| BSPRY | FLJ20150 | B-box and SPRY domain containing | - | HPRD | 10978534 |
| ENAH | ENA | MENA | NDPP1 | enabled homolog (Drosophila) | - | HPRD,BioGRID | 10801818 |
| LASP1 | Lasp-1 | MLN50 | LIM and SH3 protein 1 | Reconstituted Complex Two-hybrid | BioGRID | 15004028 |
| LATS1 | WARTS | wts | LATS, large tumor suppressor, homolog 1 (Drosophila) | Affinity Capture-Western Reconstituted Complex | BioGRID | 10831611 |
| NEBL | FLJ53769 | LNEBL | MGC119746 | MGC119747 | bA56H7.1 | nebulette | - | HPRD,BioGRID | 15004028 |
| NEDD9 | CAS-L | CAS2 | CASL | CASS2 | HEF1 | dJ49G10.2 | dJ761I2.1 | neural precursor cell expressed, developmentally down-regulated 9 | - | HPRD,BioGRID | 11782456 |
| RPH3AL | NOC2 | rabphilin 3A-like (without C2 domains) | - | HPRD | 9367993 |
| TES | DKFZp586B2022 | MGC1146 | TESS | TESS-2 | testis derived transcript (3 LIM domains) | Reconstituted Complex | BioGRID | 12695497 |
| VASP | - | vasodilator-stimulated phosphoprotein | - | HPRD | 7644520 |10801818 |10851246 |10882740 |
| VASP | - | vasodilator-stimulated phosphoprotein | Affinity Capture-Western in vitro in vivo Two-hybrid | BioGRID | 10801818 |10882740 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD | 8622875 |
| VAV3 | FLJ40431 | vav 3 guanine nucleotide exchange factor | - | HPRD | 8622875 |
| ZHX1 | - | zinc fingers and homeoboxes 1 | Two-hybrid | BioGRID | 12659632 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| PID ILK PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN KERATINOCYTE PATHWAY | 21 | 19 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR1 PATHWAY | 43 | 32 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP | 214 | 155 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR UP | 164 | 122 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS UP | 293 | 179 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN UP | 239 | 157 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 4 | 16 | 9 | All SZGR 2.0 genes in this pathway |
| WANG METHYLATED IN BREAST CANCER | 35 | 25 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| LIU LIVER CANCER | 38 | 18 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| ZUCCHI METASTASIS DN | 44 | 29 | All SZGR 2.0 genes in this pathway |
| BASSO B LYMPHOCYTE NETWORK | 143 | 96 | All SZGR 2.0 genes in this pathway |
| YAGI AML RELAPSE PROGNOSIS | 35 | 24 | All SZGR 2.0 genes in this pathway |
| GOLUB ALL VS AML DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| YAGI AML SURVIVAL | 129 | 87 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS BY TBH AND H2O2 | 36 | 24 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| MACLACHLAN BRCA1 TARGETS DN | 16 | 12 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA EARLY RESPONSE TO TGFB1 | 58 | 43 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| VERRECCHIA RESPONSE TO TGFB1 C3 | 14 | 10 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A UP | 111 | 70 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE UP | 81 | 53 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| JIANG TIP30 TARGETS DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| PARK APL PATHOGENESIS DN | 50 | 35 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE DN | 75 | 61 | All SZGR 2.0 genes in this pathway |
| KARLSSON TGFB1 TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS LOW SERUM | 100 | 51 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| WIERENGA PML INTERACTOME | 42 | 23 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-103/107 | 280 | 286 | 1A | hsa-miR-103brain | AGCAGCAUUGUACAGGGCUAUGA |
| hsa-miR-107brain | AGCAGCAUUGUACAGGGCUAUCA | ||||
| miR-15/16/195/424/497 | 268 | 274 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG | ||||
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.