Gene Page: APOL1
Summary ?
| GeneID | 8542 |
| Symbol | APOL1 |
| Synonyms | APO-L|APOL|APOL-I|FSGS4 |
| Description | apolipoprotein L1 |
| Reference | MIM:603743|HGNC:HGNC:618|Ensembl:ENSG00000100342|HPRD:04774|Vega:OTTHUMG00000030427 |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 0.255 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
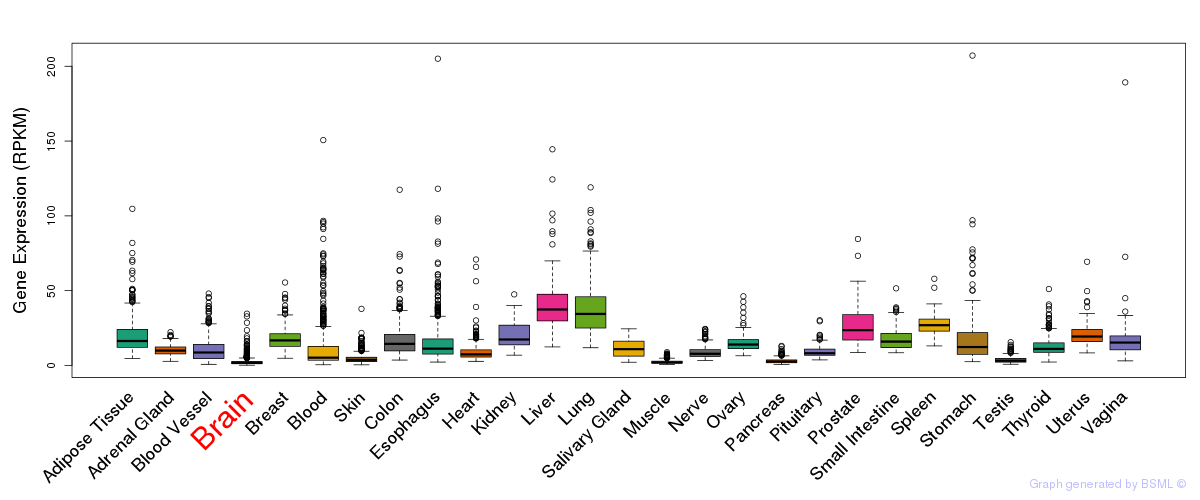
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM117B | 0.96 | 0.97 |
| AL096711.2 | 0.96 | 0.97 |
| CLASP1 | 0.96 | 0.97 |
| NCAM1 | 0.96 | 0.96 |
| BRD3 | 0.96 | 0.96 |
| ZNF445 | 0.95 | 0.97 |
| BCORL1 | 0.95 | 0.96 |
| GPR161 | 0.95 | 0.92 |
| SRGAP2 | 0.95 | 0.97 |
| KIAA1549 | 0.95 | 0.97 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.71 | -0.82 |
| AIFM3 | -0.71 | -0.81 |
| HLA-F | -0.70 | -0.79 |
| ALDOC | -0.69 | -0.74 |
| HEPN1 | -0.69 | -0.78 |
| SERPINB6 | -0.69 | -0.77 |
| TSC22D4 | -0.69 | -0.83 |
| S100B | -0.69 | -0.88 |
| FBXO2 | -0.68 | -0.66 |
| CLU | -0.68 | -0.73 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 12621437 | |
| GO:0005319 | lipid transporter activity | IEA | - | |
| GO:0005254 | chloride channel activity | IDA | 16020735 | |
| GO:0008289 | lipid binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008202 | steroid metabolic process | IEA | - | |
| GO:0008203 | cholesterol metabolic process | IEA | - | |
| GO:0019835 | cytolysis | IDA | 12621437 |17192540 | |
| GO:0006821 | chloride transport | IDA | 16020735 | |
| GO:0006629 | lipid metabolic process | IEA | - | |
| GO:0006869 | lipid transport | IEA | - | |
| GO:0031640 | killing of cells of another organism | IDA | 12621437 |17192540 | |
| GO:0042157 | lipoprotein metabolic process | IEA | - | |
| GO:0045087 | innate immune response | IDA | 17192540 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | IDA | 17192540 | |
| GO:0031224 | intrinsic to membrane | IC | 16020735 | |
| GO:0034364 | high-density lipoprotein particle | IDA | 9325276 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS DN | 637 | 377 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG UP | 78 | 50 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING UP | 83 | 56 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN UP | 30 | 21 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| CROONQUIST IL6 DEPRIVATION UP | 20 | 10 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MS DN | 46 | 24 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA1 UP | 76 | 46 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| BOSCO INTERFERON INDUCED ANTIVIRAL MODULE | 78 | 48 | All SZGR 2.0 genes in this pathway |