Gene Page: CNOT8
Summary ?
| GeneID | 9337 |
| Symbol | CNOT8 |
| Synonyms | CAF1|CALIF|Caf1b|POP2|hCAF1 |
| Description | CCR4-NOT transcription complex subunit 8 |
| Reference | MIM:603731|HGNC:HGNC:9207|Ensembl:ENSG00000155508|HPRD:07225|Vega:OTTHUMG00000130192 |
| Gene type | protein-coding |
| Map location | 5q31-q33 |
| Pascal p-value | 0.168 |
| Sherlock p-value | 0.264 |
| Fetal beta | 0.953 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cortex Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00459 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01718 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18632102 | 5 | 154238496 | CNOT8 | 4.04E-8 | -0.01 | 1.14E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17116417 | chr5 | 154145473 | CNOT8 | 9337 | 0.08 | cis | ||
| rs816718 | chr5 | 154184147 | CNOT8 | 9337 | 0.03 | cis | ||
| rs2688201 | chr5 | 154252772 | CNOT8 | 9337 | 0.08 | cis | ||
| rs17116534 | chr5 | 154254817 | CNOT8 | 9337 | 0.05 | cis | ||
| rs2544880 | chr5 | 154265118 | CNOT8 | 9337 | 0.11 | cis | ||
| rs1035368 | chr5 | 154308306 | CNOT8 | 9337 | 0.08 | cis | ||
| rs348752 | chr5 | 154324528 | CNOT8 | 9337 | 0.04 | cis | ||
| rs1561519 | chr2 | 35823020 | CNOT8 | 9337 | 0.06 | trans | ||
| rs7591339 | chr2 | 35825597 | CNOT8 | 9337 | 0.06 | trans | ||
| rs7570146 | chr2 | 35844751 | CNOT8 | 9337 | 0.06 | trans | ||
| rs1439691 | chr2 | 35851281 | CNOT8 | 9337 | 0.06 | trans | ||
| rs1944366 | chr18 | 52713130 | CNOT8 | 9337 | 0.03 | trans | ||
| rs12326929 | chr18 | 70864592 | CNOT8 | 9337 | 0.17 | trans |
Section II. Transcriptome annotation
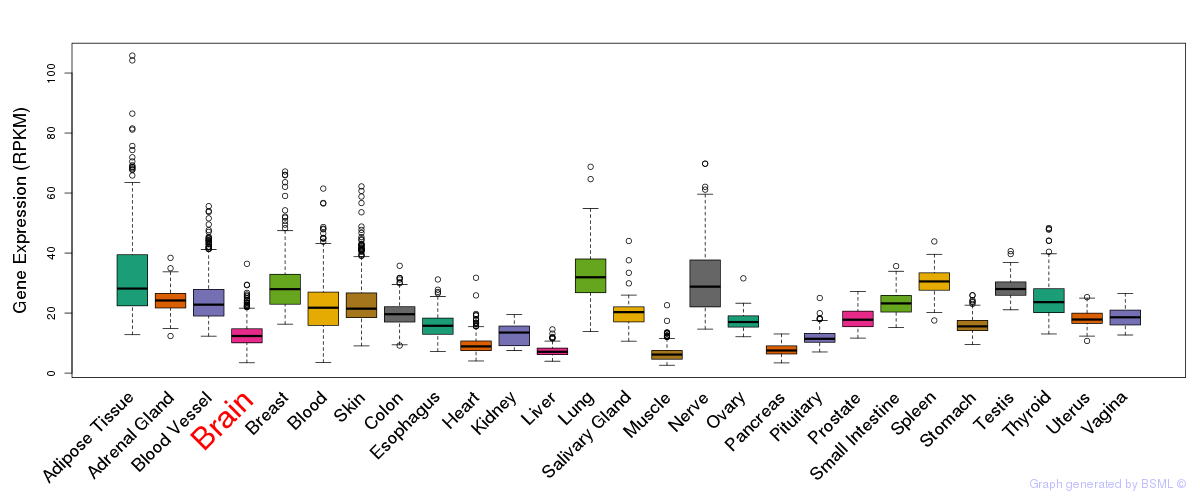
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BUB3 | 0.92 | 0.90 |
| H2AFY | 0.92 | 0.89 |
| CSTF2 | 0.92 | 0.91 |
| POLDIP3 | 0.92 | 0.91 |
| ALDH18A1 | 0.91 | 0.90 |
| USP39 | 0.91 | 0.89 |
| SFRS1 | 0.91 | 0.89 |
| TRA2B | 0.91 | 0.88 |
| RFWD3 | 0.90 | 0.90 |
| TH1L | 0.90 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.70 | -0.84 |
| MT-CO2 | -0.69 | -0.84 |
| AF347015.33 | -0.69 | -0.82 |
| AF347015.31 | -0.68 | -0.82 |
| AF347015.8 | -0.67 | -0.83 |
| FXYD1 | -0.67 | -0.80 |
| MT-CYB | -0.66 | -0.80 |
| C5orf53 | -0.66 | -0.71 |
| S100B | -0.64 | -0.75 |
| AF347015.21 | -0.64 | -0.84 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0003700 | transcription factor activity | NAS | 10036195 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | NAS | 10036195 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | TAS | 9820826 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IDA | 11256614 | |
| GO:0005634 | nucleus | NAS | 10036195 | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG RNA DEGRADATION | 59 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION OF MRNA | 22 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF MRNA | 284 | 128 | All SZGR 2.0 genes in this pathway |
| REACTOME DEADENYLATION DEPENDENT MRNA DECAY | 48 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR DN | 66 | 42 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| SEIDEN MET SIGNALING | 19 | 16 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO HD MTX DN | 24 | 18 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| CHIARETTI ACUTE LYMPHOBLASTIC LEUKEMIA ZAP70 | 67 | 33 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-138 | 259 | 266 | 1A,m8 | hsa-miR-138brain | AGCUGGUGUUGUGAAUC |
| miR-200bc/429 | 1266 | 1272 | 1A | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-7 | 209 | 215 | m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.