Gene Page: PICK1
Summary ?
| GeneID | 9463 |
| Symbol | PICK1 |
| Synonyms | PICK|PRKCABP |
| Description | protein interacting with PRKCA 1 |
| Reference | MIM:605926|HGNC:HGNC:9394|Ensembl:ENSG00000100151|HPRD:16176|Vega:OTTHUMG00000151159 |
| Gene type | protein-coding |
| Map location | 22q13.1 |
| Pascal p-value | 0.318 |
| Sherlock p-value | 0.589 |
| Fetal beta | -0.319 |
| eGene | Caudate basal ganglia Hippocampus Putamen basal ganglia Myers' cis & trans Meta |
| Support | PROTEIN CLUSTERING G2Cdb.humanPSD G2Cdb.humanPSP Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 2.7105 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6596223 | chr5 | 134774309 | PICK1 | 9463 | 0.14 | trans | ||
| rs11912649 | 22 | 38415542 | PICK1 | ENSG00000100151.11 | 2.422E-6 | 0.02 | -36776 | gtex_brain_putamen_basal |
| rs1883471 | 22 | 38417510 | PICK1 | ENSG00000100151.11 | 1.929E-6 | 0.02 | -34808 | gtex_brain_putamen_basal |
| rs5995530 | 22 | 38419738 | PICK1 | ENSG00000100151.11 | 2.622E-6 | 0.02 | -32580 | gtex_brain_putamen_basal |
| rs713991 | 22 | 38426043 | PICK1 | ENSG00000100151.11 | 1.922E-6 | 0.02 | -26275 | gtex_brain_putamen_basal |
| rs3026670 | 22 | 38426774 | PICK1 | ENSG00000100151.11 | 1.64E-6 | 0.02 | -25544 | gtex_brain_putamen_basal |
| rs10427673 | 22 | 38427551 | PICK1 | ENSG00000100151.11 | 1.891E-6 | 0.02 | -24767 | gtex_brain_putamen_basal |
| rs8142174 | 22 | 38427963 | PICK1 | ENSG00000100151.11 | 1.561E-6 | 0.02 | -24355 | gtex_brain_putamen_basal |
| rs6519095 | 22 | 38431917 | PICK1 | ENSG00000100151.11 | 1.551E-6 | 0.02 | -20401 | gtex_brain_putamen_basal |
| rs12157609 | 22 | 38432277 | PICK1 | ENSG00000100151.11 | 1.242E-6 | 0.02 | -20041 | gtex_brain_putamen_basal |
| rs4821730 | 22 | 38441587 | PICK1 | ENSG00000100151.11 | 8.873E-7 | 0.02 | -10731 | gtex_brain_putamen_basal |
| rs4821731 | 22 | 38445214 | PICK1 | ENSG00000100151.11 | 9.399E-7 | 0.02 | -7104 | gtex_brain_putamen_basal |
| rs5756890 | 22 | 38445400 | PICK1 | ENSG00000100151.11 | 1.083E-6 | 0.02 | -6918 | gtex_brain_putamen_basal |
| rs760973 | 22 | 38447266 | PICK1 | ENSG00000100151.11 | 9.684E-7 | 0.02 | -5052 | gtex_brain_putamen_basal |
| rs4384887 | 22 | 38447365 | PICK1 | ENSG00000100151.11 | 1.12E-6 | 0.02 | -4953 | gtex_brain_putamen_basal |
| rs742395 | 22 | 38447444 | PICK1 | ENSG00000100151.11 | 9.21E-7 | 0.02 | -4874 | gtex_brain_putamen_basal |
| rs2413493 | 22 | 38448295 | PICK1 | ENSG00000100151.11 | 1.424E-6 | 0.02 | -4023 | gtex_brain_putamen_basal |
| rs4821733 | 22 | 38449394 | PICK1 | ENSG00000100151.11 | 2.354E-6 | 0.02 | -2924 | gtex_brain_putamen_basal |
| . | 22 | 38449820 | PICK1 | ENSG00000100151.11 | 2.179E-6 | 0.02 | -2498 | gtex_brain_putamen_basal |
| rs5756894 | 22 | 38450136 | PICK1 | ENSG00000100151.11 | 2.332E-6 | 0.02 | -2182 | gtex_brain_putamen_basal |
| rs2018980 | 22 | 38450620 | PICK1 | ENSG00000100151.11 | 1.693E-6 | 0.02 | -1698 | gtex_brain_putamen_basal |
| rs742396 | 22 | 38452254 | PICK1 | ENSG00000100151.11 | 9.904E-7 | 0.02 | -64 | gtex_brain_putamen_basal |
| rs738442 | 22 | 38457329 | PICK1 | ENSG00000100151.11 | 2.578E-6 | 0.02 | 5011 | gtex_brain_putamen_basal |
| rs738443 | 22 | 38458681 | PICK1 | ENSG00000100151.11 | 9.904E-7 | 0.02 | 6363 | gtex_brain_putamen_basal |
| rs4821735 | 22 | 38458969 | PICK1 | ENSG00000100151.11 | 9.904E-7 | 0.02 | 6651 | gtex_brain_putamen_basal |
| rs6000991 | 22 | 38460484 | PICK1 | ENSG00000100151.11 | 9.904E-7 | 0.02 | 8166 | gtex_brain_putamen_basal |
| rs2076369 | 22 | 38463652 | PICK1 | ENSG00000100151.11 | 9.904E-7 | 0.02 | 11334 | gtex_brain_putamen_basal |
| rs2076370 | 22 | 38463968 | PICK1 | ENSG00000100151.11 | 9.904E-7 | 0.02 | 11650 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
General gene expression (GTEx)
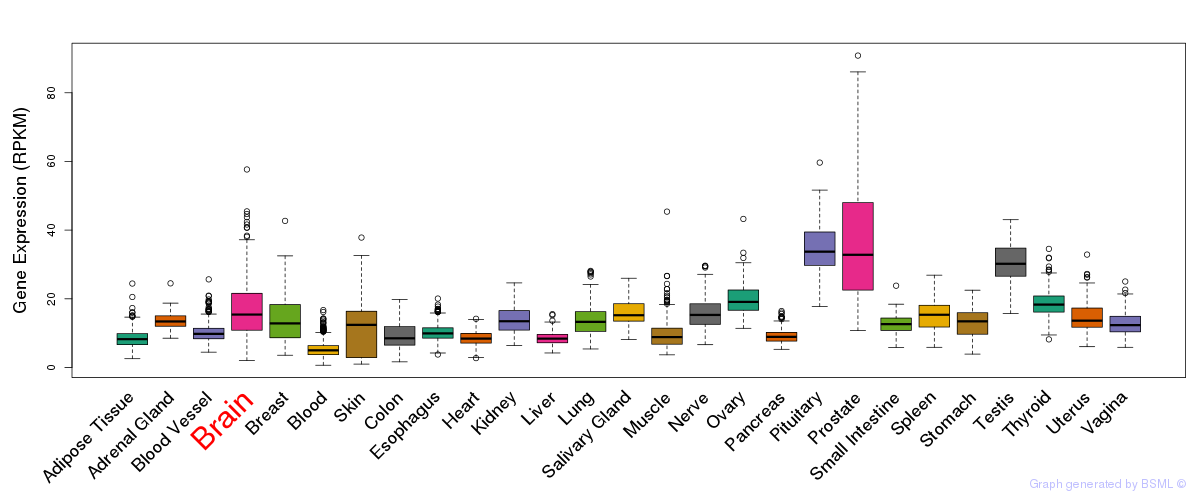
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CPNE6 | 0.60 | 0.68 |
| NELF | 0.58 | 0.65 |
| PRKCG | 0.57 | 0.68 |
| CD6 | 0.57 | 0.57 |
| CNGB1 | 0.57 | 0.60 |
| FIBCD1 | 0.56 | 0.50 |
| SUSD4 | 0.56 | 0.61 |
| SYT17 | 0.55 | 0.62 |
| GALNTL4 | 0.54 | 0.53 |
| TTC7B | 0.54 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1949 | -0.27 | -0.22 |
| RBMX2 | -0.26 | -0.39 |
| ZNF193 | -0.25 | -0.18 |
| FAM167B | -0.25 | -0.39 |
| TUBB2B | -0.25 | -0.26 |
| SH2B2 | -0.25 | -0.34 |
| AP000349.1 | -0.25 | -0.27 |
| SPATA9 | -0.25 | -0.26 |
| TMSB15A | -0.25 | -0.28 |
| ZNF300 | -0.25 | -0.13 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | ISS | Neurotransmitter (GO term level: 4) | - |
| GO:0005080 | protein kinase C binding | ISS | - | |
| GO:0008022 | protein C-terminus binding | IPI | 11343649 | |
| GO:0016887 | ATPase activity | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045161 | neuronal ion channel clustering | TAS | neuron (GO term level: 11) | 11343649 |
| GO:0007205 | activation of protein kinase C activity | IDA | 10623590 | |
| GO:0006468 | protein amino acid phosphorylation | ISS | - | |
| GO:0006605 | protein targeting | IEA | - | |
| GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | NAS | 10623590 | |
| GO:0015844 | monoamine transport | IDA | 11343649 | |
| GO:0043046 | DNA methylation during gametogenesis | TAS | 12138111 | |
| GO:0043113 | receptor clustering | ISS | - | |
| GO:0043045 | DNA methylation during embryonic development | NAS | 12138111 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042734 | presynaptic membrane | IDA | neuron, axon, Synap (GO term level: 5) | 11343649 |
| GO:0045202 | synapse | ISS | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005794 | Golgi apparatus | ISS | - | |
| GO:0005737 | cytoplasm | IDA | 11343649 | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 11343649 | |
| GO:0048471 | perinuclear region of cytoplasm | ISS | - | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACCN1 | ACCN | ASIC2 | ASIC2a | BNC1 | BNaC1 | MDEG | hBNaC1 | amiloride-sensitive cation channel 1, neuronal | PICK1 interacts with BNC1a. | BIND | 11802773 |
| ACCN1 | ACCN | ASIC2 | ASIC2a | BNC1 | BNaC1 | MDEG | hBNaC1 | amiloride-sensitive cation channel 1, neuronal | - | HPRD | 11739374 |
| ACCN1 | ACCN | ASIC2 | ASIC2a | BNC1 | BNaC1 | MDEG | hBNaC1 | amiloride-sensitive cation channel 1, neuronal | PICK1 interacts with BNC1b. | BIND | 11802773 |
| ACCN2 | ASIC | ASIC1 | ASIC1A | BNaC2 | hBNaC2 | amiloride-sensitive cation channel 2, neuronal | PICK1 interacts with ASIC. | BIND | 11802773 |
| ACCN2 | ASIC | ASIC1 | ASIC1A | BNaC2 | hBNaC2 | amiloride-sensitive cation channel 2, neuronal | - | HPRD,BioGRID | 11739374 |11802773 |12578970 |
| BNC1 | BNC | BSN1 | HsT19447 | basonuclin 1 | Affinity Capture-Western Two-hybrid | BioGRID | 11802773 |
| BTG2 | MGC126063 | MGC126064 | PC3 | TIS21 | BTG family, member 2 | - | HPRD | 11237868 |
| EFNB1 | CFND | CFNS | EFL3 | EPLG2 | Elk-L | LERK2 | MGC8782 | ephrin-B1 | - | HPRD,BioGRID | 9883737 |
| ERBB2 | CD340 | HER-2 | HER-2/neu | HER2 | NEU | NGL | TKR1 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian) | - | HPRD,BioGRID | 11278603 |
| ERBB2IP | ERBIN | LAP2 | erbb2 interacting protein | Two-hybrid | BioGRID | 11278603 |
| GRIA1 | GLUH1 | GLUR1 | GLURA | HBGR1 | MGC133252 | glutamate receptor, ionotropic, AMPA 1 | - | HPRD,BioGRID | 11891216 |
| GRIA2 | GLUR2 | GLURB | GluR-K2 | HBGR2 | glutamate receptor, ionotropic, AMPA 2 | Reconstituted Complex Two-hybrid | BioGRID | 11891216 |
| GRIA3 | GLUR-C | GLUR-K3 | GLUR3 | GLURC | MRX94 | glutamate receptor, ionotrophic, AMPA 3 | Reconstituted Complex Two-hybrid | BioGRID | 11891216 |
| GRIA4 | GLUR4 | GLUR4C | GLURD | glutamate receptor, ionotrophic, AMPA 4 | Reconstituted Complex Two-hybrid | BioGRID | 11891216 |
| GRIK1 | EAA3 | EEA3 | GLR5 | GLUR5 | glutamate receptor, ionotropic, kainate 1 | - | HPRD | 11891216 |
| GRIK1 | EAA3 | EEA3 | GLR5 | GLUR5 | glutamate receptor, ionotropic, kainate 1 | - | HPRD,BioGRID | 12597860 |
| GRIK2 | EAA4 | GLR6 | GLUK6 | GLUR6 | MGC74427 | MRT6 | glutamate receptor, ionotropic, kainate 2 | - | HPRD,BioGRID | 12597860 |
| GRIK3 | EAA5 | GLR7 | GLUR7 | GluR7a | glutamate receptor, ionotropic, kainate 3 | Reconstituted Complex Two-hybrid | BioGRID | 11891216 |
| GRM3 | GLUR3 | GPRC1C | MGLUR3 | mGlu3 | glutamate receptor, metabotropic 3 | Reconstituted Complex Two-hybrid | BioGRID | 11891216 |
| GRM7 | FLJ40498 | GLUR7 | GPRC1G | MGLUR7 | mGlu7 | glutamate receptor, metabotropic 7 | PICK1 interacts with mGluR7b. This interaction was modeled on a demonstrated interaction between PICK1 from an unspecified species and human mGluR7b. | BIND | 11891216 |
| GRM7 | FLJ40498 | GLUR7 | GPRC1G | MGLUR7 | mGlu7 | glutamate receptor, metabotropic 7 | Affinity Capture-Western Biochemical Activity Reconstituted Complex Two-hybrid | BioGRID | 11007882 |
| LRP2BP | DKFZp761O0113 | FLJ44965 | LRP2 binding protein | - | HPRD | 12508107 |
| PICK1 | MGC15204 | PICK | PRKCABP | protein interacting with PRKCA 1 | PICK1 interacts with another PICK1. | BIND | 11802773 |
| PRKCA | AAG6 | MGC129900 | MGC129901 | PKC-alpha | PKCA | PRKACA | protein kinase C, alpha | Affinity Capture-Western | BioGRID | 11007882 |
| PRLHR | GPR10 | GR3 | MGC126539 | MGC126541 | PrRPR | prolactin releasing hormone receptor | Affinity Capture-Western | BioGRID | 11641419 |
| SLC6A3 | DAT | DAT1 | solute carrier family 6 (neurotransmitter transporter, dopamine), member 3 | - | HPRD,BioGRID | 11343649 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF AMPA RECEPTORS | 28 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | 16 | 11 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| PENG LEUCINE DEPRIVATION UP | 142 | 93 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| MAGRANGEAS MULTIPLE MYELOMA IGLL VS IGLK UP | 42 | 24 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| MOREAUX B LYMPHOCYTE MATURATION BY TACI UP | 92 | 58 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS UP | 69 | 41 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| MCCABE HOXC6 TARGETS CANCER UP | 31 | 23 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA DN | 68 | 45 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA DN | 80 | 53 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-214 | 96 | 102 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.