Gene Page: MTRF1
Summary ?
| GeneID | 9617 |
| Symbol | MTRF1 |
| Synonyms | MRF1|MTTRF1|RF1 |
| Description | mitochondrial translational release factor 1 |
| Reference | MIM:604601|HGNC:HGNC:7469|Ensembl:ENSG00000120662|HPRD:06855|Vega:OTTHUMG00000016790 |
| Gene type | protein-coding |
| Map location | 13q14.1-q14.3 |
| Pascal p-value | 0.013 |
| Sherlock p-value | 0.302 |
| Fetal beta | 0.527 |
| eGene | Cerebellar Hemisphere Cerebellum Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17012924 | chr2 | 33612474 | MTRF1 | 9617 | 0.1 | trans | ||
| rs870686 | chr8 | 25253521 | MTRF1 | 9617 | 0.15 | trans | ||
| rs10521998 | chrX | 32437852 | MTRF1 | 9617 | 0.13 | trans | ||
| rs16998308 | chrX | 32442345 | MTRF1 | 9617 | 0.03 | trans |
Section II. Transcriptome annotation
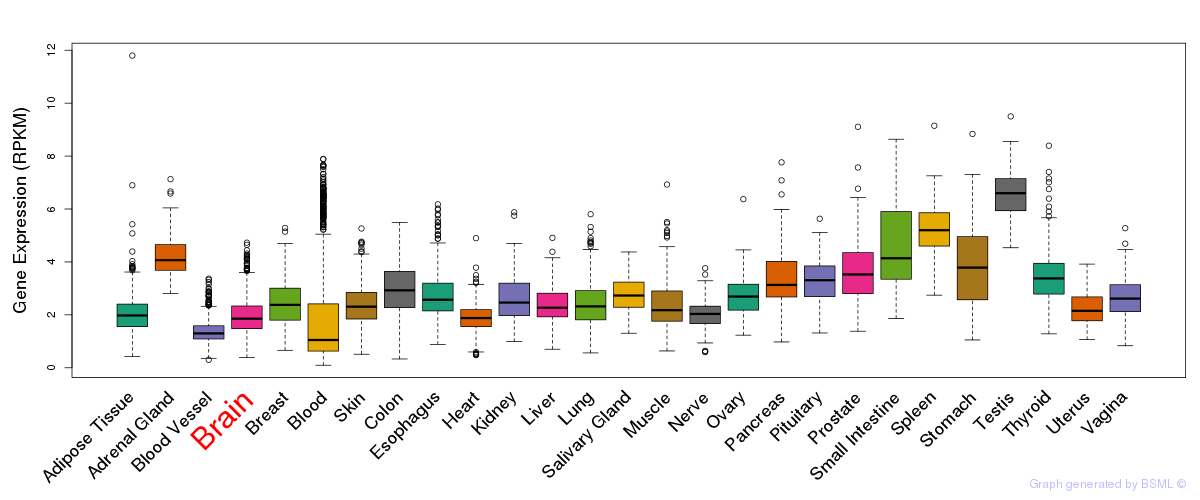
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRBA | 0.83 | 0.84 |
| SPAG9 | 0.81 | 0.83 |
| LONP2 | 0.80 | 0.83 |
| PDZD8 | 0.80 | 0.83 |
| IPO8 | 0.80 | 0.80 |
| WDFY1 | 0.80 | 0.81 |
| SCRN1 | 0.80 | 0.81 |
| ACBD5 | 0.80 | 0.79 |
| GZF1 | 0.80 | 0.82 |
| LRRC58 | 0.80 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.59 | -0.54 |
| AF347015.31 | -0.51 | -0.52 |
| MT-CO2 | -0.51 | -0.51 |
| C1orf54 | -0.50 | -0.49 |
| AF347015.8 | -0.50 | -0.49 |
| AF347015.27 | -0.49 | -0.49 |
| MYL3 | -0.48 | -0.48 |
| SYCP3 | -0.48 | -0.46 |
| IL32 | -0.47 | -0.44 |
| FXYD1 | -0.47 | -0.45 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016149 | translation release factor activity, codon specific | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006449 | regulation of translational termination | TAS | 9838146 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | TAS | 9838146 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| MANTOVANI NFKB TARGETS UP | 43 | 33 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS EARLY PROGENITOR | 532 | 309 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |