| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 6477 |
Name | SIAH1 |
Synonymous | siah E3 ubiquitin protein ligase 1;SIAH1;siah E3 ubiquitin protein ligase 1 |
Definition | E3 ubiquitin-protein ligase SIAH1|seven in absentia homolog 1|siah-1a |
Position | 16q12.1 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
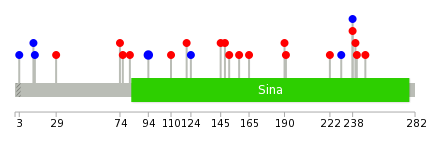 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
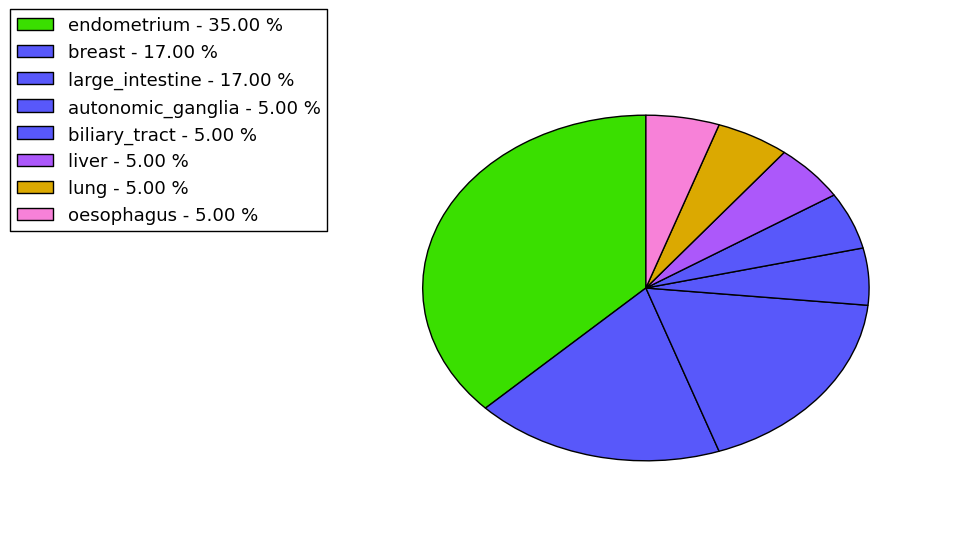 |
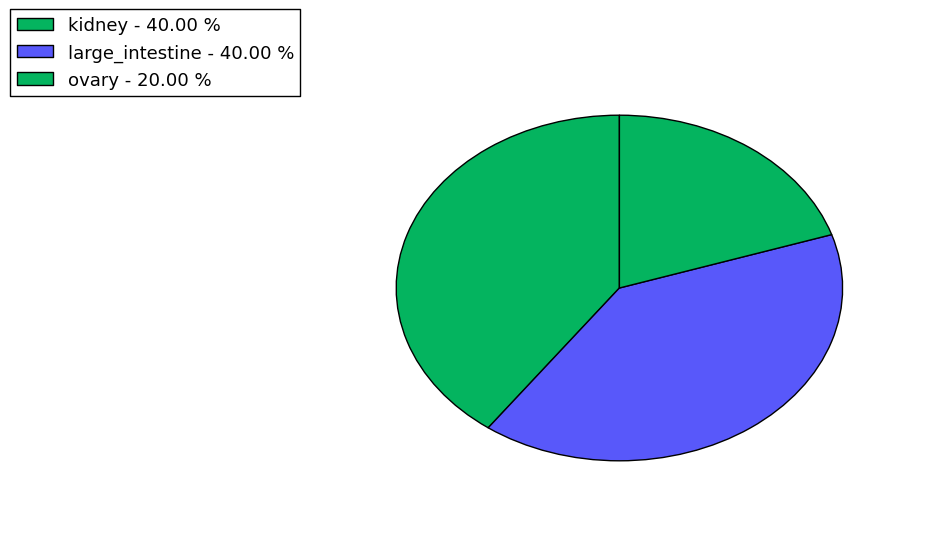 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.06. |
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.714G>A; p.A238A; 16:48361715-48361715 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.231C>T; p.G77G; 16:48362198-48362198 |
large_intestine | carcinoma; adenocarcinoma | Substitution - coding silent |
c.231C>T; p.G77G; 16:48362198-48362198 |
large_intestine | carcinoma; adenocarcinoma | Substitution - coding silent |
c.718C>T; p.P240S; 16:48361711-48361711 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.361T>A; p.C121S; 16:48362068-48362068 |
skin | malignant_melanoma | Substitution - Missense |
c.242C>A; p.S81Y; 16:48362187-48362187 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.370A>C; p.R124R; 16:48362059-48362059 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.600T>C; p.D200D; 16:48361829-48361829 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.451C>G; p.Q151E; 16:48361978-48361978 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.741T>G; p.I247M; 16:48361688-48361688 |
liver | carcinoma; hepatocellular_carcinoma | Substitution - Missense |
c.226C>T; p.R76W; 16:48362203-48362203 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.713C>T; p.A238V; 16:48361716-48361716 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.443T>A; p.L148Q; 16:48361986-48361986 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.9T>C; p.R3R; 16:48362420-48362420 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.693G>A; p.R231R; 16:48361736-48361736 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.494T>C; p.F165S; 16:48361935-48361935 |
autonomic_ganglia | neuroblastoma | Substitution - Missense |
c.39C>G; p.T13T; 16:48362390-48362390 |
ovary | carcinoma; serous_carcinoma | Substitution - coding silent |
c.10C>T; p.Q4*; 16:48362419-48362419 |
breast | carcinoma | Substitution - Nonsense |
c.472C>A; p.L158I; 16:48361957-48361957 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.221C>T; p.T74I; 16:48362208-48362208 |
oesophagus | carcinoma; adenocarcinoma | Substitution - Missense |
c.664G>A; p.A222T; 16:48361765-48361765 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.85A>G; p.T29A; 16:48362344-48362344 |
breast | carcinoma | Substitution - Missense |
c.435G>A; p.M145I; 16:48361994-48361994 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.570G>A; p.M190I; 16:48361859-48361859 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.42G>A; p.S14S; 16:48362387-48362387 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.690T>C; p.H230H; 16:48361739-48361739 |
thyroid | carcinoma | Substitution - coding silent |
c.571T>G; p.L191V; 16:48361858-48361858 |
biliary_tract; gallbladder | carcinoma; adenocarcinoma | Substitution - Missense |
c.722G>A; p.R241Q; 16:48361707-48361707 |
breast | carcinoma | Substitution - Missense |
c.514C>G; p.L172V; 16:48361915-48361915 |
oesophagus; lower_third | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.282A>T; p.V94V; 16:48362147-48362147 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - coding silent |
c.282A>T; p.V94V; 16:48362147-48362147 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - coding silent |
c.328C>A; p.P110T; 16:48362101-48362101 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |